Batch Multiple Simulations Run
This RMarkdown file executes run type of batch simulation. The script utilizes the gastroPlusAPI package to communicate with the GastroPlus X 10.2 service. A batch simulation allows you to run multiple defined simulations for any number of compounds or scenarios.
The output from the batch simulation run is then used to create a Cp-time plot.
To customize your study with this script for a different simulation / project / variables, please make changes to the set-input-information code-chunk.
Configure required packages
Load other necessary packages(tidyverse) required to execute the data manipulation and visualization in the script
Load gastroPlusAPI package
Load gastroPlusRModuLens package
library(tidyverse)
library(gastroPlusAPI)
library(gastroPlusRModuLens)
# set default ggplot2 theme and options
theme_set(
theme_gastroPlus_grid()
)
set_default_ggplot_options()Set working directory
Set working directory as the current source editor context
if (rstudioapi::isAvailable()){
current_working_directory <- dirname(rstudioapi::getSourceEditorContext()$path)
setwd(current_working_directory)
}Start GPX Service
Establishes a connection to the GastroPlus service, which allows R to communicate with GastroPlus through its API
gpx_service <- start_service(verbose=FALSE)✔ Configured the GastroPlus Servicegpx_service$is_alive()[1] TRUESetup Input Information
Make modification to the variables in this chunk to customize your workflow.
project_path: Location of the gpx project
simulation_name: Subset of simulation names to include in the run, or NULL to include all simulations
run_name: Name of the run that will be created
project_path <- "../../ProjectFiles/GPX Library.gpproject"
simulation_name <- NULL
run_name <- "BatchSimsRun"Execute the Run
Based on the details mentioned in the previous code chunk, the batch simulation run will be created, executed and results will be shown
#Load the project
open_project(project_path)
if (is.null(simulation_name)) {
simulation_name <- get_simulations()$simulation_name
}
#Create run and execute simulations
create_run(run_name = run_name, run_type = RunType$BatchSimulation)
add_simulations_to_run(run_name = run_name, simulation_names = simulation_name)
execute_run(run_name = run_name)Process Run Output
This section retrieves and processes the results from the batch simulation run, including:
Getting summary output with pharmacokinetic parameters
Identifying and retrieving the concentration-time series data
Retrieving observed data for comparison
Build Series Descriptors
Series descriptors include the compound name and description of what is measured. Some series descriptors are defined by only the compound name and some StateType. Other series descriptors are defined by additional features, like CompartmentType.
The build_series_descriptors() function accepts one or more compound names along with one or more unnamed sets of series descriptors (without the compound name). For series descriptors that are defined by multiple features - like CompartmentType and StateType, wrap in c().
all_compounds <- unlist(get_project_assets("Compound")$assets)
all_compounds[1] "Atenolol" "Propranolol HCl" "Ranitidine"
[4] "Brick Dust" "Piroxicam" "Carbamazepine"
[7] "Ketoprofen" "Furosemide" "Metoprolol Tartrate"All series descriptors could be generated with one call to build_series_descriptors(), or certain series could grouped together for data manipulation purposes after collecting the data.
cp_series_descriptors <- build_series_descriptors(
compound = all_compounds,
c(CompartmentType$SystemicCirculation, StateType$ConcentrationPlasma),
# Alternative: will check for venous return concentration (PBPK model)
c(CompartmentType$VenousReturn, StateType$ConcentrationPlasma)
)
mass_series_descriptors <- build_series_descriptors(
compound = all_compounds,
StateType$TotalAbsorbed,
StateType$TotalDissolved
)
series_descriptors <- c(
cp_series_descriptors,
mass_series_descriptors
)Get Simulated Data
The functions get_series_data_project(), get_series_data_run(), and get_series_data_simulation() are all extensions of gastroPlusAPI::get_series_data() that operate on multiple runs, simulations, and series.
get_series_data_project()collects the desired series for the entire project, optionally subset to some values ofrun_nameand/orsimulation_name.get_series_data_run()collects the desired series for one run, optionally subset to some values ofsimulation_name.get_series_data_simulation()collects the desired series for one simulation in one run.
series_data <- get_series_data_run(
run_name = run_name,
series_descriptor = series_descriptors
)
glimpse(series_data)Rows: 1,387
Columns: 10
$ run_name <chr> "BatchSimsRun", "BatchSimsRun", "Batch…
$ simulation_name <chr> "Atenolol 100mg PO tablet", "Atenolol …
$ series_name <chr> "Atenolol - Systemic Circulation - Con…
$ dependent_unit <chr> "ng/mL", "ng/mL", "ng/mL", "ng/mL", "n…
$ independent_unit <chr> "h", "h", "h", "h", "h", "h", "h", "h"…
$ iteration <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ dependent <dbl> 0.000000e+00, 2.651325e-16, 4.043352e-…
$ independent <dbl> 0.000000e+00, 2.217896e-06, 4.131786e-…
$ simulation_iteration_key_name <chr> "Atenolol 100mg PO tablet \a0", "Ateno…
$ simulation_iteration_key_name_2 <chr> "Atenolol 100mg PO tablet", "Atenolol …series_data %>%
count(series_name, independent_unit, dependent_unit)# A tibble: 27 × 4
series_name independent_unit dependent_unit n
<chr> <chr> <chr> <int>
1 Atenolol - Systemic Circulation - Conc… h ng/mL 50
2 Atenolol - Total Absorbed h mg 50
3 Atenolol - Total Dissolved h mg 50
4 Brick Dust - Systemic Circulation - Co… h ng/mL 28
5 Brick Dust - Total Absorbed h mg 28
6 Brick Dust - Total Dissolved h mg 28
7 Carbamazepine - Systemic Circulation -… h ng/mL 49
8 Carbamazepine - Total Absorbed h mg 49
9 Carbamazepine - Total Dissolved h mg 49
10 Furosemide - Systemic Circulation - Co… h ng/mL 60
# ℹ 17 more rowsseries_data %>%
count(run_name, simulation_name)# A tibble: 9 × 3
run_name simulation_name n
<chr> <chr> <int>
1 BatchSimsRun Atenolol 100mg PO tablet 150
2 BatchSimsRun Brick Dust 10mg PO tablet 84
3 BatchSimsRun Carbamazepine 200mg PO tablet 147
4 BatchSimsRun Furosemide 40mg PO tablet 180
5 BatchSimsRun Ketoprofen 100mg PO tablet 171
6 BatchSimsRun Metoprolol Tartrate 200mg PO tablet 179
7 BatchSimsRun Piroxicam 20mg PO capsule 143
8 BatchSimsRun Propranolol HCl 160mg PO tablet 174
9 BatchSimsRun Ranitidine 150mg PO tablet 159Get Observed Data
The get_observed_series_data() function is an extension of gastroPlusAPI::get_observed_series() that operates on any subset of group_name, group_type, series_name, and/or series_type and always returns a data frame. When called with no arguments, get_observed_series_data() collects all observed data in the open project. This function also calculates standard deviation (std_dev) derived from reported mean and CV%.
observed_data_inventory <- get_experimental_data_inventory()
observed_data_inventory# A tibble: 6 × 4
group_name group_type series_name series_type
<chr> <chr> <chr> <chr>
1 Ketoprofen Solubility Solubility Solubility vs pH pH_Solubil…
2 Propranolol HCl Solubility Solubility Solubility vs pH pH_Solubil…
3 Piroxicam Solubility Solubility Solubility vs pH pH_Solubil…
4 Piroxicam PO Cp time ExposureData Piroxicam Exposure Pl… UncertainC…
5 Propranolol HCl PO Cp time ExposureData Propranolol HCl Expos… UncertainC…
6 Metoprolol Tartrate PO Cp time ExposureData Metoprolol Tartrate E… UncertainC…observed_data <- get_observed_series_data()
observed_exposure_data <- observed_data %>%
filter(group_type == "ExposureData")
observed_solubility_data <- observed_data %>%
filter(group_type == "Solubility")
glimpse(observed_exposure_data)Rows: 30
Columns: 20
$ group_name <chr> "Piroxicam PO Cp time", "Piroxicam PO Cp time",…
$ group_type <chr> "ExposureData", "ExposureData", "ExposureData",…
$ series_name <chr> "Piroxicam Exposure Plasma", "Piroxicam Exposur…
$ series_type <chr> "UncertainConcentrationSeries", "UncertainConce…
$ independent_unit <chr> "h", "h", "h", "h", "h", "h", "h", "h", "h", "h…
$ dependent_unit <chr> "ng/mL", "ng/mL", "ng/mL", "ng/mL", "ng/mL", "n…
$ independent <dbl> 0.0, 0.5, 1.0, 2.0, 3.0, 6.0, 12.0, 24.0, 48.0,…
$ dependent <dbl> 0.0, 31.4, 933.9, 2507.6, 2609.4, 2112.4, 1774.…
$ module <chr> "SystemicCirculation", "SystemicCirculation", "…
$ section <chr> "", "", "", "", "", "", "", "", "", "", "", "",…
$ compartment <chr> "SystemicCirculation", "SystemicCirculation", "…
$ state <chr> "ConcentrationPlasma", "ConcentrationPlasma", "…
$ dose <dbl> 0.0200, 0.0200, 0.0200, 0.0200, 0.0200, 0.0200,…
$ dose_unit <chr> "g", "g", "g", "g", "g", "g", "g", "g", "g", "g…
$ infusion_time <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
$ infusion_time_unit <chr> "s", "s", "s", "s", "s", "s", "s", "s", "s", "s…
$ body_mass <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 74000, 7…
$ body_mass_unit <chr> "g", "g", "g", "g", "g", "g", "g", "g", "g", "g…
$ uncertainty_percentage <dbl> 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.…
$ std_dev <dbl> 0.0000, 0.0000, 0.0000, 0.0000, 0.0000, 0.0000,…observed_exposure_data %>%
count(series_name, series_type)# A tibble: 3 × 3
series_name series_type n
<chr> <chr> <int>
1 Metoprolol Tartrate Exposure Plasma UncertainConcentrationSeries 9
2 Piroxicam Exposure Plasma UncertainConcentrationSeries 13
3 Propranolol HCl Exposure Plasma UncertainConcentrationSeries 8Plot Results
Define common values across simulated and observed data
Since the names of runs, simulations, and observed series are user-defined, the shared variable names in simulated and observed data will not always match and may require some data manipulation.
In many projects, even though both series_data and observed_data include the series_name variable, but the content of this variable is not always well aligned.
intersect(names(series_data), names(observed_exposure_data))[1] "series_name" "dependent_unit" "independent_unit" "dependent"
[5] "independent" series_data %>%
count(series_name, independent_unit, dependent_unit)# A tibble: 27 × 4
series_name independent_unit dependent_unit n
<chr> <chr> <chr> <int>
1 Atenolol - Systemic Circulation - Conc… h ng/mL 50
2 Atenolol - Total Absorbed h mg 50
3 Atenolol - Total Dissolved h mg 50
4 Brick Dust - Systemic Circulation - Co… h ng/mL 28
5 Brick Dust - Total Absorbed h mg 28
6 Brick Dust - Total Dissolved h mg 28
7 Carbamazepine - Systemic Circulation -… h ng/mL 49
8 Carbamazepine - Total Absorbed h mg 49
9 Carbamazepine - Total Dissolved h mg 49
10 Furosemide - Systemic Circulation - Co… h ng/mL 60
# ℹ 17 more rowsobserved_exposure_data %>%
count(series_name, independent_unit, dependent_unit)# A tibble: 3 × 4
series_name independent_unit dependent_unit n
<chr> <chr> <chr> <int>
1 Metoprolol Tartrate Exposure Plasma h ng/mL 9
2 Piroxicam Exposure Plasma h ng/mL 13
3 Propranolol HCl Exposure Plasma h ng/mL 8The desired content to align simulated and observed data might exist in other variables.
setdiff(names(series_data), names(observed_exposure_data))[1] "run_name" "simulation_name"
[3] "iteration" "simulation_iteration_key_name"
[5] "simulation_iteration_key_name_2"series_data %>%
count(run_name, simulation_name)# A tibble: 9 × 3
run_name simulation_name n
<chr> <chr> <int>
1 BatchSimsRun Atenolol 100mg PO tablet 150
2 BatchSimsRun Brick Dust 10mg PO tablet 84
3 BatchSimsRun Carbamazepine 200mg PO tablet 147
4 BatchSimsRun Furosemide 40mg PO tablet 180
5 BatchSimsRun Ketoprofen 100mg PO tablet 171
6 BatchSimsRun Metoprolol Tartrate 200mg PO tablet 179
7 BatchSimsRun Piroxicam 20mg PO capsule 143
8 BatchSimsRun Propranolol HCl 160mg PO tablet 174
9 BatchSimsRun Ranitidine 150mg PO tablet 159setdiff(names(observed_exposure_data), names(series_data)) [1] "group_name" "group_type" "series_type"
[4] "module" "section" "compartment"
[7] "state" "dose" "dose_unit"
[10] "infusion_time" "infusion_time_unit" "body_mass"
[13] "body_mass_unit" "uncertainty_percentage" "std_dev" observed_exposure_data %>%
count(series_type)# A tibble: 1 × 2
series_type n
<chr> <int>
1 UncertainConcentrationSeries 30Aligning the most important features of simulated and observed data will produce the most meaningful visualizations.
series_data <- series_data %>%
mutate(
category = case_when(
series_name %in% cp_series_descriptors ~ "Concentration",
series_name %in% mass_series_descriptors ~ "Mass"
),
endpoint = str_remove(series_name, "^[^-]*-\\s?"),
compound = trimws(str_replace(series_name, "(^[^-]*)?-.*", "\\1"))
)
series_data %>%
count(category, endpoint, compound)# A tibble: 27 × 4
category endpoint compound n
<chr> <chr> <chr> <int>
1 Concentration Systemic Circulation - Concentration Plasma Atenolol 50
2 Concentration Systemic Circulation - Concentration Plasma Brick Dust 28
3 Concentration Systemic Circulation - Concentration Plasma Carbamazepine 49
4 Concentration Systemic Circulation - Concentration Plasma Furosemide 60
5 Concentration Systemic Circulation - Concentration Plasma Ketoprofen 57
6 Concentration Systemic Circulation - Concentration Plasma Metoprolol T… 65
7 Concentration Systemic Circulation - Concentration Plasma Piroxicam 55
8 Concentration Systemic Circulation - Concentration Plasma Propranolol … 62
9 Concentration Systemic Circulation - Concentration Plasma Ranitidine 53
10 Mass Total Absorbed Atenolol 50
# ℹ 17 more rowsobserved_exposure_data <- observed_exposure_data %>%
mutate(
category = "Concentration",
endpoint = "Systemic Circulation - Concentration Plasma",
compound = trimws(str_remove(series_name, "Exposure Plasma"))
)
observed_exposure_data %>%
count(category, endpoint, compound, dose, dose_unit)# A tibble: 3 × 6
category endpoint compound dose dose_unit n
<chr> <chr> <chr> <dbl> <chr> <int>
1 Concentration Systemic Circulation - Concentra… Metopro… 0.156 g 9
2 Concentration Systemic Circulation - Concentra… Piroxic… 0.02 g 13
3 Concentration Systemic Circulation - Concentra… Propran… 0.140 g 8# subset of simulated data including concentration data.
cp_series_data <- series_data %>%
filter(
category == "Concentration"
)
cp_series_data %>%
count(category, endpoint, compound)# A tibble: 9 × 4
category endpoint compound n
<chr> <chr> <chr> <int>
1 Concentration Systemic Circulation - Concentration Plasma Atenolol 50
2 Concentration Systemic Circulation - Concentration Plasma Brick Dust 28
3 Concentration Systemic Circulation - Concentration Plasma Carbamazepine 49
4 Concentration Systemic Circulation - Concentration Plasma Furosemide 60
5 Concentration Systemic Circulation - Concentration Plasma Ketoprofen 57
6 Concentration Systemic Circulation - Concentration Plasma Metoprolol Ta… 65
7 Concentration Systemic Circulation - Concentration Plasma Piroxicam 55
8 Concentration Systemic Circulation - Concentration Plasma Propranolol H… 62
9 Concentration Systemic Circulation - Concentration Plasma Ranitidine 53Concentration vs Time
cp_plot_list <- unique(cp_series_data$compound) %>%
map(function(.compound) {
cp_series_subset <- cp_series_data %>%
filter(compound == .compound)
observed_exposure_subset <- observed_exposure_data %>%
filter(compound == .compound)
plot_series_data(
data_sim = cp_series_subset,
data_obs = observed_exposure_subset,
x_label = "Time (h)",
y_label = "Concentration (ng/mL)",
) +
labs(
subtitle = .compound
)
})
ggpubr::ggarrange(plotlist = cp_plot_list)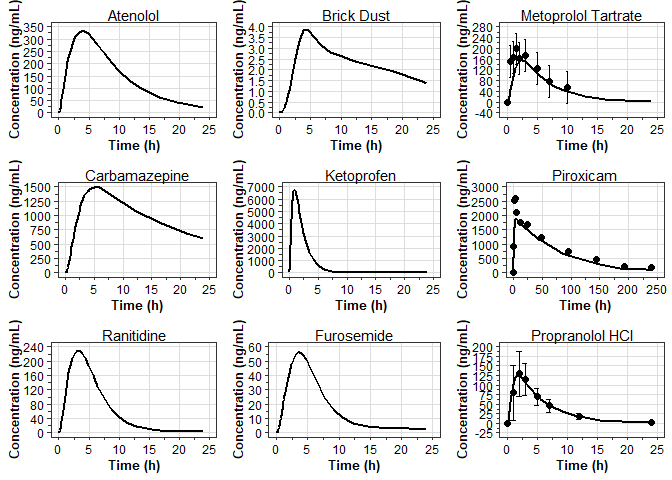
Concentration and Mass vs Time
When data with 2 unique values of dependent_unit is provided to plot_series_data(), a dual-axis is created.
# concentration and mass-time curves (dual axis).
cp_mass_plot_list <- unique(series_data$compound) %>%
map(function(.compound) {
series_subset <- series_data %>%
filter(compound == .compound)
plot_series_data(
series_subset,
color = endpoint,
linetype = category,
x_label = "Time (h)",
y_label = "Concentration\n(ng/mL)",
y2_label = "Mass (mg)"
) +
labs(
subtitle = .compound,
color = NULL,
linetype = NULL
) +
guides(
color = guide_legend(NULL)
)
})
ggpubr::ggarrange(plotlist = cp_mass_plot_list, common.legend = TRUE)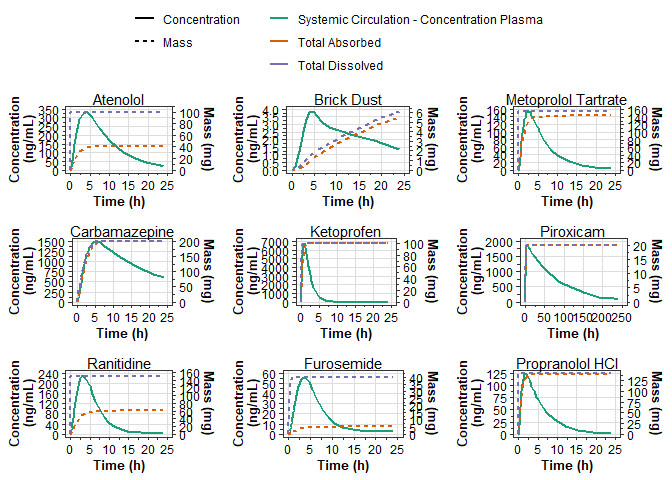
Solubility Observed Data
plot_series_data(
data_obs = observed_solubility_data,
x_label = "pH",
y_label = "Solubility (ng/mL)",
) +
scale_y_continuous() +
facet_wrap(~ group_name, scales = "free_y")Scale for y is already present.
Adding another scale for y, which will replace the existing scale.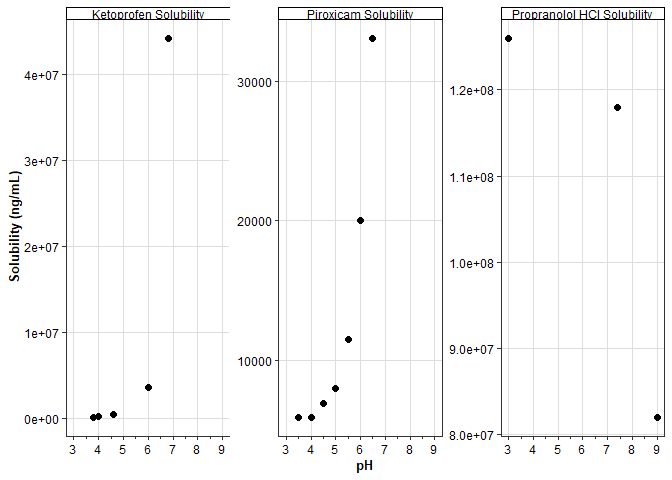
Diagnostic plots
With observed and predicted data available, we can also plot observed vs predicted values or residuals vs predicted values.
Observed vs Predicted
intersect(names(observed_exposure_data), names(cp_series_data))[1] "series_name" "independent_unit" "dependent_unit" "independent"
[5] "dependent" "category" "endpoint" "compound" # combine observed and simulated data.
data_obs_pred <- cp_series_data %>%
select(-c(series_name)) %>%
rename(Predicted = dependent) %>%
# inner join to only keep the rows with matching by variables.
inner_join(
observed_exposure_data %>%
select(-c(series_name)) %>%
rename(Observed = dependent),
by = join_by(
independent_unit, dependent_unit, independent, endpoint, category, compound
)
) %>%
mutate(
Residuals = Observed - Predicted
)
plot_obs_vs_pred(
data = data_obs_pred,
obs = Observed,
pred = Predicted,
) +
facet_wrap(~ compound)Warning in scale_x_continuous(..., transform = transform_log10()): log-10
transformation introduced infinite values.Warning in scale_y_continuous(..., transform = transform_log10()): log-10
transformation introduced infinite values.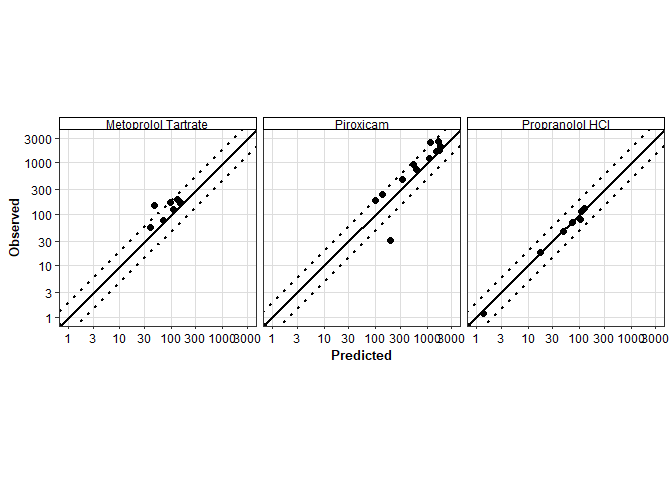
Residuals vs Predicted
plot_res_vs_pred(
data = data_obs_pred,
res = Residuals,
pred = Predicted
) +
scale_x_continuous() +
scale_y_continuous() +
facet_wrap(~ compound, scales = "free")Scale for x is already present.
Adding another scale for x, which will replace the existing scale.
Scale for y is already present.
Adding another scale for y, which will replace the existing scale.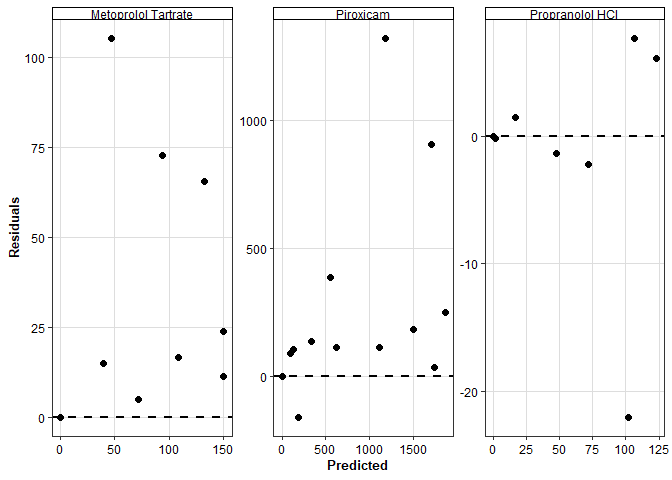
Kill GPX Service
gpx_service$kill()[1] TRUE