Sequential and Cross PSA Run Mode
This RMarkdown file creates and executes a Sequential or Cross PSA run. The script utilizes the gastroPlusAPI package to communicate with the GastroPlus X 10.2 service.
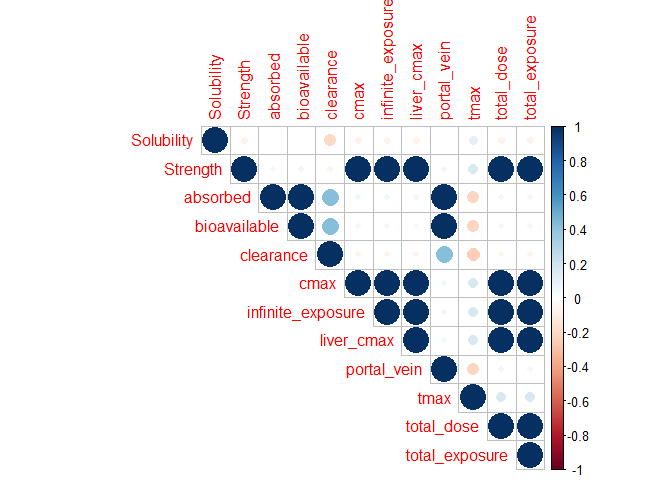
The output from the PSA run is then used to create a 3D plot between two input variables and one output variable and a correlation plot between the input variables and the pharmacokinetic output variables.
To customize your study with this script for a different simulation / project / variables, please make changes to the “Set Input Information” section.
Configure required packages
Load other necessary packages required to execute the script
Load gastroPlusAPI package
Load gastroPlusRModuLens package
library(tidyverse)
library(gastroPlusAPI)
library(gastroPlusRModuLens)Set working directory
Set working directory as the current source editor context
if (rstudioapi::isAvailable()){
current_working_directory <- dirname(rstudioapi::getSourceEditorContext()$path)
setwd(current_working_directory)
}Start GPX Service
gpx_service <- start_service(verbose=FALSE)✔ Configured the GastroPlus Servicegpx_service$is_alive()[1] TRUESetup Input Information
Make modification to the variables in this chunk to customize your analysis.
project_path = Location of the project where PSA run will be created
simulation_name = Name of the simulation for the PSA run
psa_analysis_mode = PSA Analysis mode (Sequential / Cross)
run_name = Name of the PSA run that will be created
input_variables_tibble = Tibble containing the variable information required to setup the PSA run.
The specified user-defined variable values will be used in each simulation of the PSA run.
project_path = "../../ProjectFiles/GPX Library.gpproject"
simulation_name = "Atenolol 100mg PO tablet"
psa_analysis_mode = ParameterSensitivityAnalysisMode$Sequential
run_name = "sequential_psa"
input_variables_tibble <- tibble(variable_name = c("Strength", "Solubility"),
spacing = c("Logarithmic", "Uniform"),
lower_bound = c(50, 10000),
upper_bound = c(500, 50000),
variable_unit = c("milligram", "gram/meters cubed"),
number_of_tests = c(10,5))
#Variables for 3d plot - choose from any input variable for PSA and summary output variables
variables_x_y_z_3d_plot = c(x= "Strength", y="Solubility", z="cmax")Configure and Execute the PSA Run
Based on the variables mentioned in the previous code chunk, the PSA run will be configured and executed
#Load the project
open_project(project_path)
#Create new PSA run
create_run(run_name, RunType$ParameterSensitivityAnalysis)
add_simulations_to_run(run_name, c(simulation_name))
variable_information <- get_variable_information(run_name,
simulation_name,
input_variables_tibble$variable_name)
#Setup PSA Parameters List
psa_parameter_list <- vector(mode="list",length=nrow(input_variables_tibble))
count = 1
for(var in input_variables_tibble$variable_name){
var_info <- variable_information %>% filter(input_variable_name == var)
input_info <- input_variables_tibble %>% filter(variable_name == var)
psa_parameter_list[[count]] <- PSAParameter$new(
variable_name = var,
baseline_value = var_info$variable_value$value,
lower_bound = input_info$lower_bound,
upper_bound = input_info$upper_bound,
unit = input_info$variable_unit,
scalar_type = var_info$scalar_type,
number_of_tests = input_info$number_of_tests,
spacing = input_info$spacing
)
count = count + 1
}
#Setup PSA Setting
psa_setting <- PSASetting$new(
analysis_mode = psa_analysis_mode,
simulate_baseline = FALSE,
update_Kp = FALSE)
#Create PSA Configuration object
PSA_configuration <- PSAConfiguration$new(PSA_Setting = psa_setting,
PSA_Parameters = psa_parameter_list)
#Create Run Configuration object
run_configuration <- RunConfiguration$new(PSA_Configuration = PSA_configuration)
#Configure the run in GPX
configure_run(run_name, run_configuration, use_full_variable_name = FALSE)
#Execute Run
execute_run(run_name)Process run output
Get summary output and iteration records from the run just executed and create a tibble that can be used for visualization of results.
psa_data <- get_summary_output_tidy(run_name = run_name)
glimpse(psa_data)Rows: 150
Columns: 13
$ compound <chr> "Atenolol", "Atenolol", "Atenolol", "A…
$ run_simulation_iteration_key <chr> "sequential_psa - Atenolol 100mg PO ta…
$ simulation_iteration_key_name <chr> "Atenolol 100mg PO tablet | Dose: 50 m…
$ simulation_iteration_key_name_2 <chr> "Atenolol 100mg PO tablet | Dose: 50 m…
$ iteration <int> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1,…
$ name <chr> "absorbed", "bioavailable", "clearance…
$ value <dbl> 0.4105343, 0.3991967, 27.6856569, 165.…
$ unit <chr> NA, NA, "L/h", "ng/mL", "(ng/mL)*h", "…
$ label <chr> "Absorbed", "Bioavailable", "Clearance…
$ Solubility <dbl> 27000, 27000, 27000, 27000, 27000, 270…
$ Strength <dbl> 0.05000000, 0.05000000, 0.05000000, 0.…
$ Solubility_unit <chr> "g/m³", "g/m³", "g/m³", "g/m³", "g/m³"…
$ Strength_unit <chr> "g", "g", "g", "g", "g", "g", "g", "g"…#Output parameters from the PSA
psa_data %>%
count(name, unit, label)# A tibble: 10 × 4
name unit label n
<chr> <chr> <chr> <int>
1 absorbed <NA> "Absorbed" 15
2 bioavailable <NA> "Bioavailable" 15
3 clearance L/h "Clearance (L/h)" 15
4 cmax ng/mL "Cmax (ng/mL)" 15
5 infinite_exposure (ng/mL)*h "Infinite Exposure\n((ng/mL)*h)" 15
6 liver_cmax ng/mL "Liver Cmax (ng/mL)" 15
7 portal_vein <NA> "Portal Vein" 15
8 tmax h "Tmax (h)" 15
9 total_dose mg "Total Dose (mg)" 15
10 total_exposure (ng/mL)*h "Total Exposure\n((ng/mL)*h)" 15Plot results in 3D plot
Derive x, y, and z data for the 3D plot from the psa_data tibble created in the previous step.
x_unit <- unique(psa_data[[paste0(variables_x_y_z_3d_plot[["x"]], "_unit")]])
y_unit <- unique(psa_data[[paste0(variables_x_y_z_3d_plot[["y"]], "_unit")]])
psa_data_subset <- psa_data %>%
filter(name == variables_x_y_z_3d_plot[["z"]]) %>%
rename(!!variables_x_y_z_3d_plot[["z"]] := value)
z_title <- unique(psa_data_subset$label)
# 3D surface plot for Cross PSA or 3D scatter plot for other PSA
if (psa_analysis_mode == ParameterSensitivityAnalysisMode$Cross) {
fig <- plot_3d_surface(
psa_data_subset,
x = variables_x_y_z_3d_plot[["x"]],
y = variables_x_y_z_3d_plot[["y"]],
z = variables_x_y_z_3d_plot[["z"]]
)
} else {
fig <- plotly::plot_ly(
x = psa_data_subset[[variables_x_y_z_3d_plot[["x"]]]],
y = psa_data_subset[[variables_x_y_z_3d_plot[["y"]]]],
z = psa_data_subset[[variables_x_y_z_3d_plot[["z"]]]],
type = "scatter3d",
mode = "markers"
) %>%
plotly::layout(
scene = list(
xaxis = list(title = paste0(variables_x_y_z_3d_plot[["x"]], " (", x_unit, ")")),
yaxis = list(title = paste0(variables_x_y_z_3d_plot[["y"]], " (", y_unit, ")")),
zaxis = list(title = z_title)
)
)
}This is a static image of the plot, in RStudio and html, one can rotate, zoom, pan, etc.
# Within RStudio and rendering to html, the following statement shows the interactive plot
# fig
# create a png static snapshot of the plot and display the resulting png
static_plot_image <- "psa_cross_3D_plot.png"
plotly::save_image(fig, static_plot_image)
knitr::include_graphics(static_plot_image)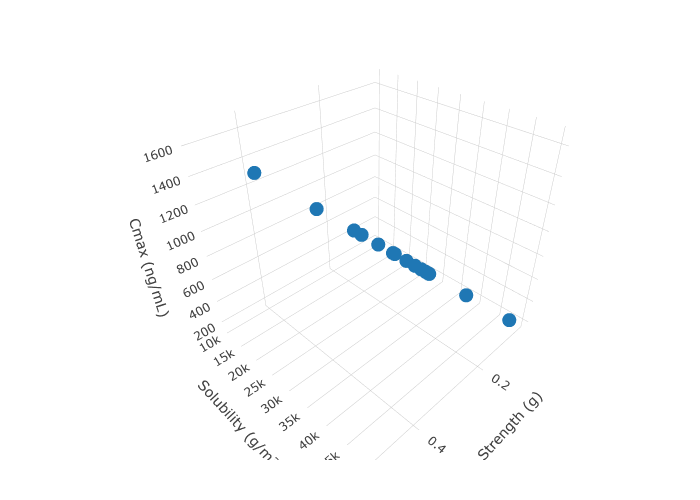
Correlation plot
cor_matrix <- psa_data %>%
select(simulation_iteration_key_name, name, where(is.numeric)) %>%
select(-any_of(c("iteration"))) %>%
pivot_wider(
names_from = name,
values_from = value
) %>%
column_to_rownames("simulation_iteration_key_name") %>%
as.matrix() %>%
cor(use = "complete.obs")
corrplot::corrplot(
corr = cor_matrix,
type = "upper"
)
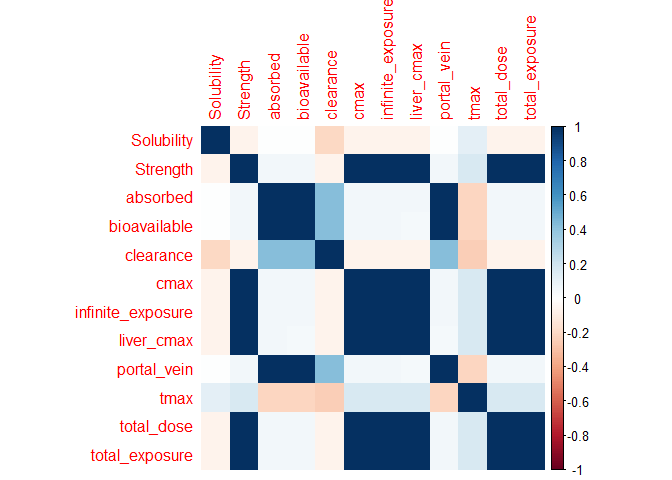
corrplot::corrplot(
corr = cor_matrix,
method = "color"
)
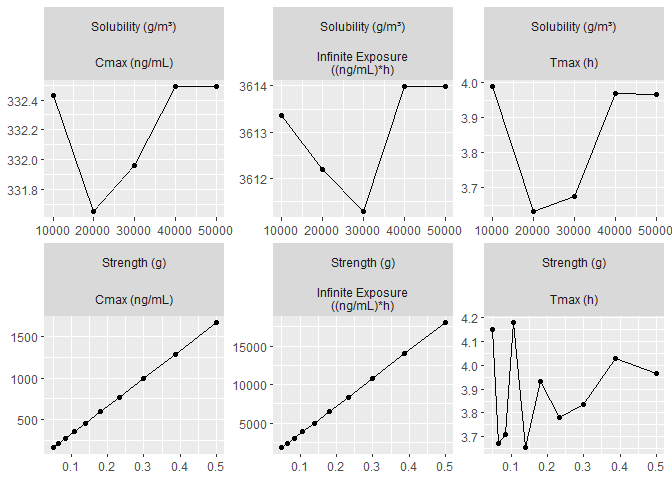
Spider plot
baseline_values <- psa_parameter_list %>%
map_dbl(~ .x$baseline_value) %>%
set_names(map_chr(psa_parameter_list, ~ .x$variable_name))
baseline_values Strength Solubility
100 27000 # amount to multiply original baseline values by to convert to output units.
unit_adjust <- psa_parameter_list %>%
map_dbl(function(.x) {
input_lower_bound <- .x$lower_bound
output_lower_bound <- min(psa_data[[.x$variable_name]])
output_lower_bound / input_lower_bound
}) %>%
set_names(map_chr(psa_parameter_list, ~ .x$variable_name))
unit_adjust Strength Solubility
0.001 1.000 baseline_values_adj <- baseline_values * unit_adjust[names(baseline_values)]
baseline_values_adj Strength Solubility
0.1 27000.0 # obtain the closest value to the baseline values.
baseline_values_closest <- baseline_values_adj %>%
imap_dbl(function(.x, .idx) {
values <- psa_data[[.idx]]
values[which.min(abs(values - .x))]
}) %>%
set_names(map_chr(psa_parameter_list, ~ .x$variable_name))
baseline_values_closest Strength Solubility
0.1 27000.0 # Use the `spider_slice()` function to subset PSA data to suitable format for a
# spider plot.
#
# For a sequential PSA, no subset is expected, but the additional variables
# generated by this function are expected for the spider plot.
psa_spider_data <- psa_data %>%
spider_slice(
baseline_values_closest
)
psa_spider_data %>%
count(parameter_name, across(any_of(names(baseline_values))))# A tibble: 15 × 4
parameter_name Strength Solubility n
<chr> <dbl> <dbl> <int>
1 Solubility 0.1 10000 10
2 Solubility 0.1 20000 10
3 Solubility 0.1 30000 10
4 Solubility 0.1 40000 10
5 Solubility 0.1 50000 10
6 Strength 0.05 27000 10
7 Strength 0.0646 27000 10
8 Strength 0.0834 27000 10
9 Strength 0.108 27000 10
10 Strength 0.139 27000 10
11 Strength 0.180 27000 10
12 Strength 0.232 27000 10
13 Strength 0.300 27000 10
14 Strength 0.387 27000 10
15 Strength 0.5 27000 10# Subset to select parameters.
psa_spider_data_subset <- psa_spider_data %>%
filter(name %in% c("cmax", "infinite_exposure", "tmax"))
plot_spider_plot(
psa_spider_data_subset
)
Delete assets and Kill GPX Service
Delete the assets created and then kill the GPX service after the script is executed
delete_run(run_name)
save_project()
gpx_service$kill()[1] TRUE