Tutorial: ACATPlus™
ACATPlus™ application
ACATPlus™ extends the ACAT model to include extra colon segments and gut wall tissue to make it more suitable for simulations of local drug disposition. ACATPlus™ adds extra colon segments, including descending, transverse, sigmoid colon, and rectum. It also includes a mucus layer and multiple gut tissue layers (mucosa, submucosa, and muscularis propria) in addition to the enterocyte layer. It could aid in the understanding of local drug disposition in the gut tissue following oral administration.
Lumen metabolism via the microbiome
Open GPX™ and, in the Dashboard view, click on the icon next to Select to open an Existing project.
Click Browse and navigate to the C:\Users\<user>\AppData\Local\Simulations Plus, Inc\GastroPlus\10.2\Tutorials\ACATPlus folder and select the project Microbiome.gpproject by clicking on it and clicking open.
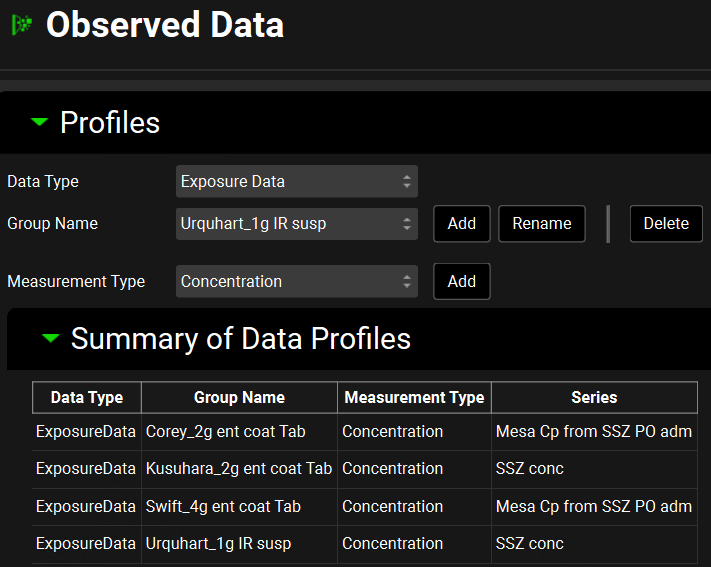
Click on the Observed Data view in the navigation pane and then review the Exposure Data. This project contains Exposure Data series of plasma concentration versus time profiles for both mesalamine and sulfasalazine compounds.
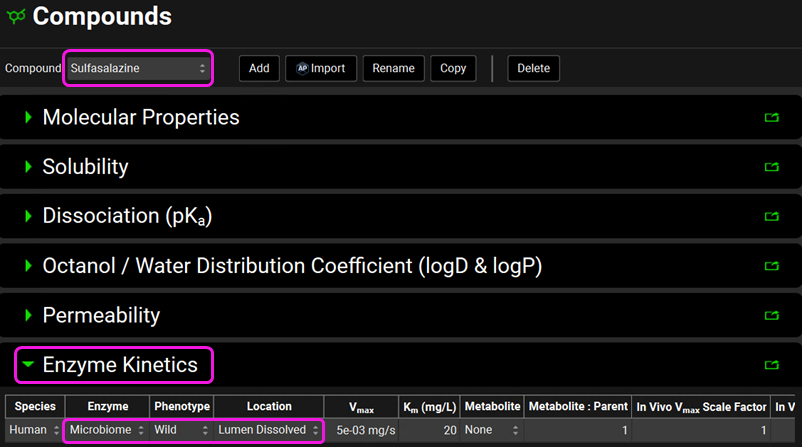
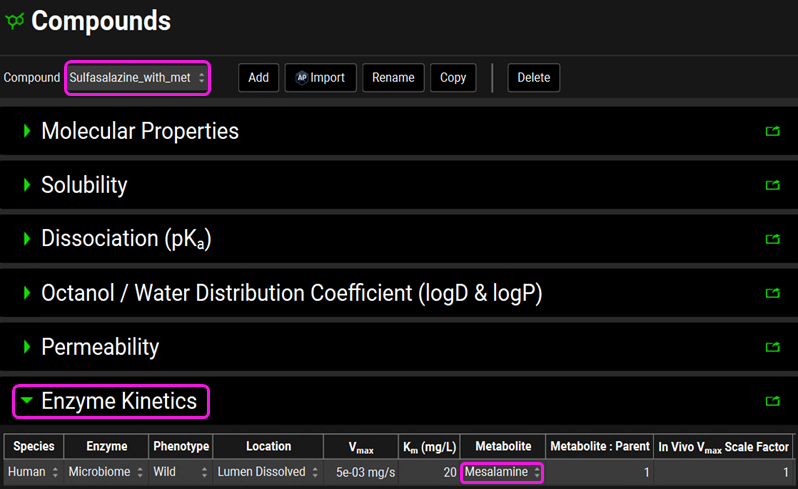
Click on the Compounds view in the navigation pane and review the inputs in each panel for Sulfasalazine. Note that sulfasalazine is a lipophilic zwitterion (logP = 3.15; one acidic pKa = 4.114, and one basic pKa = 0.55) with low solubility and permeability. The microbiome metabolism has been incorporated under the Enzyme Kinetics panel. In the gut lumen, sulfasalazine will be metabolized by the microbiome.
Click on the Dosing view in the navigation pane and review Formulations and Dosing Schedules. Two formulations are included: IR suspension, and enteric-coated tablet (PO ent coated Tab).
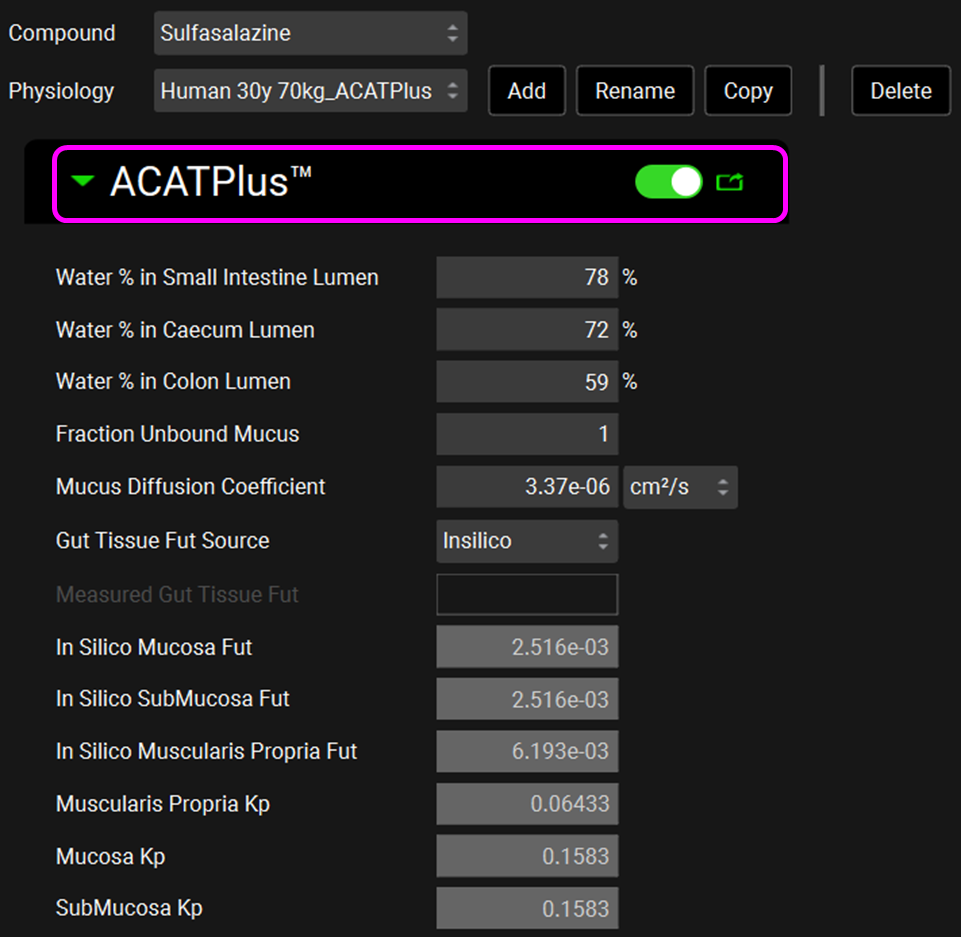
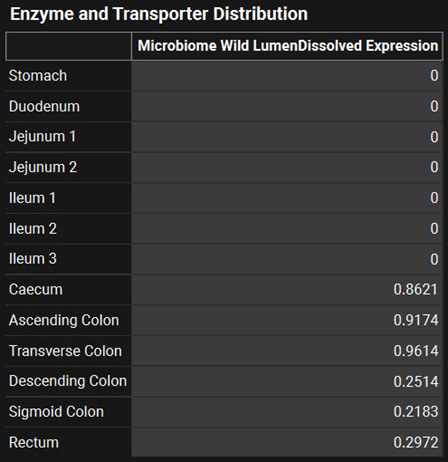
Click on the Physiologies view in the navigation pane and confirm that there are physiologies only for humans. Check the physiologies “Human 30y 70kg_ACATPlus” and “Human 34y 70kg_ACATPlus” when the compound Sulfasalazine is selected. For these 2 physiologies, the ACATPlus™ Model has been toggled on in the Gastrointestinal subpanel. Scroll down under the Gastrointestinal panel to find the distribution of microbiome in the gut lumen. Notice there is no expression in the small intestine, only in the colon. Leave the physiology set to “Human 30y 70kg_ACATPlus”.
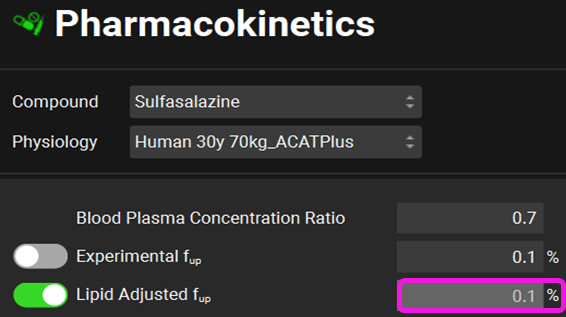
Click on the Pharmacokinetics view in the navigation pane. The Experimental Fup being very low means an extensive binding to albumin. This extensive binding does not allow the drug to bind to lipids which is why the lipid Adjusted Fup is estimated to also be at 0.1%.
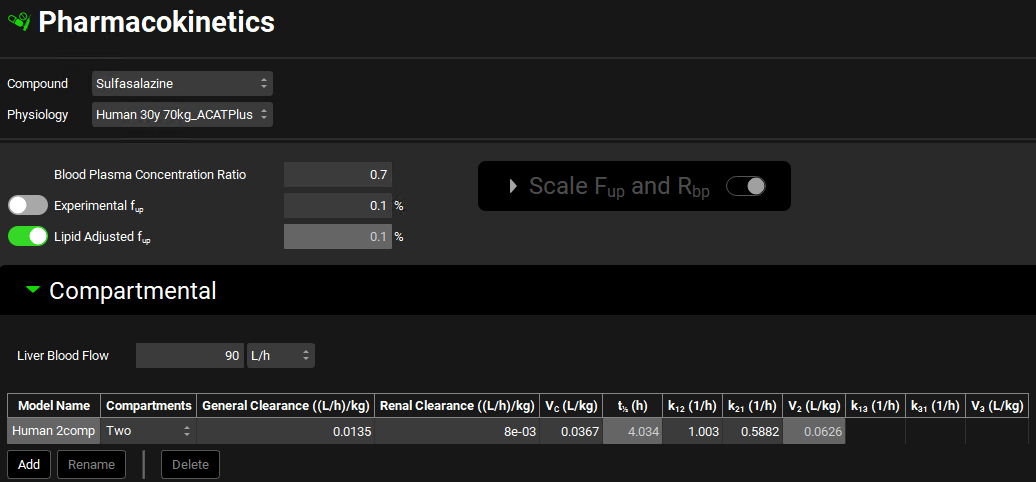
Expand the Compartmental panel by clicking on the green arrow or double-clicking on the panel header bar, and review the model. A two-compartment model is available. This two-compartment model has been created and validated based on preclinical IV data.
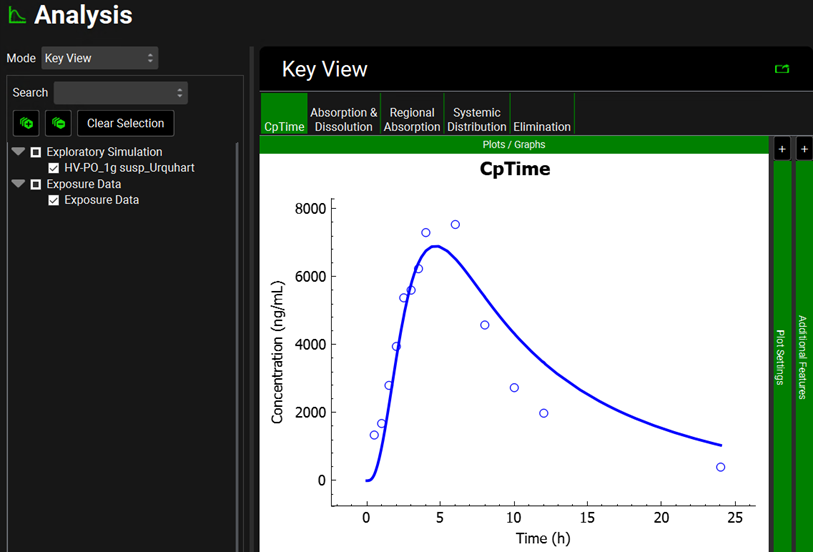
Click on the Simulations view in the navigation pane and then click on Run Simulation for “HV-PO_1g susp_Urquhart”.
Once in the Analysis view, you will see that the simulated Cp Time matches the observed data pretty well for a single dose of 1000 mg of sulfasalazine administered to healthy subjects as a suspension.
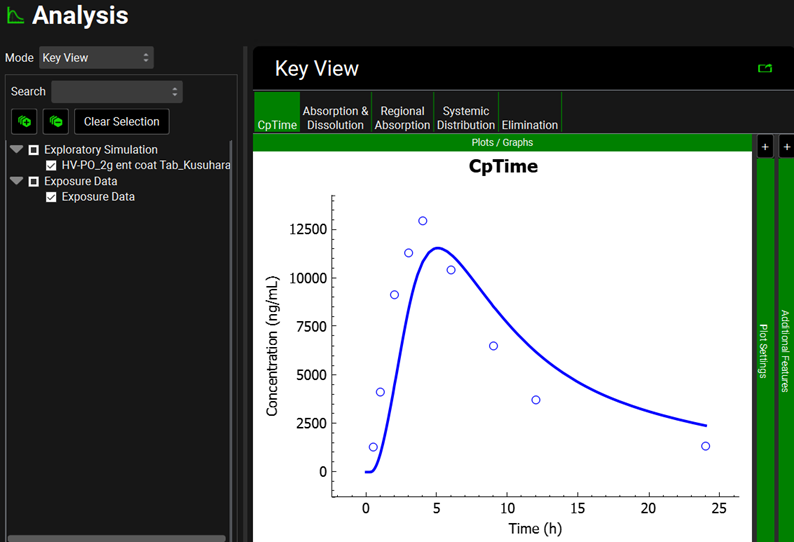
Click again on the Simulations view in the navigation pane and then click on Run Simulation for “HV-PO_2g ent coat Tab_Kusuhara”.
Once in the Analysis view, you will see that the simulated Cp Time is reasonable compared to the observed data for a single dose of 2000 mg of sulfasalazine administered to healthy subjects as an enteric-coated tablet.
Click on the Compounds view in the navigation pane and select the “Sulfasalazine_with_met” compound. It has the same input as the Sulfasalazine compound, the only difference is that the mesalamine is here linked to the sulfasalazine as its metabolite. This means that the microbiome metabolizes the sulfasalazine into mesalamine in the gut lumen.
You will now simulate both sulfasalazine and mesalamine Cp time profiles. Click on the Simulations view in the navigation pane and then click on Run Simulation for “HV-PO_2g ent coat Tab_Corey”. Because those simulations contain ACATPlus™, the time for the simulation to complete can take longer than usual based on your computer power.
Once in the Analysis view, you will see the simulated CpTime curve for mesalamine and sulfasalazine for a single dose of 2000 mg of sulfasalazine administered to healthy subjects as an enteric-coated tablet. The second screenshot represents the CpTime curves on a semi-logarithmic scale.
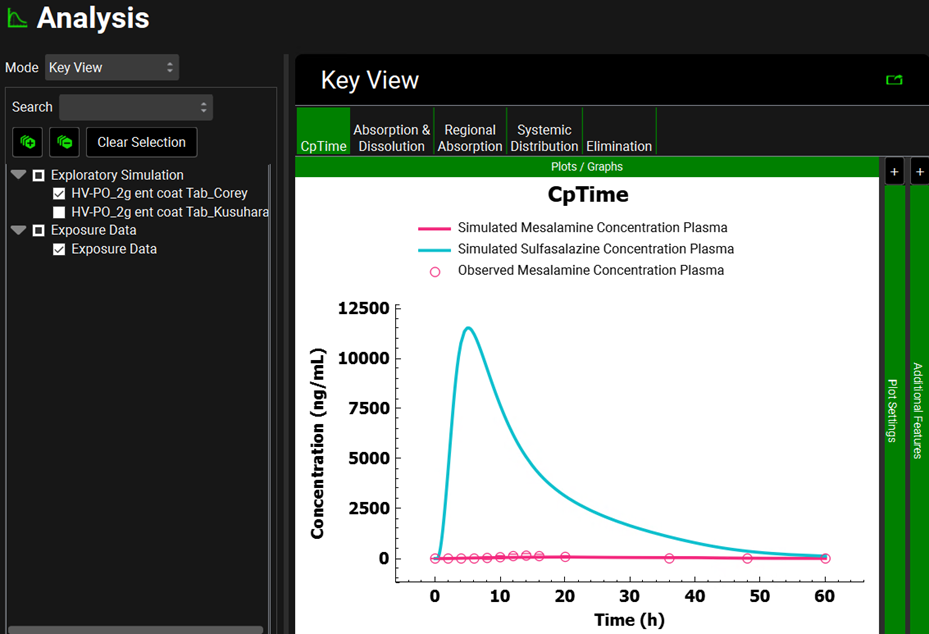
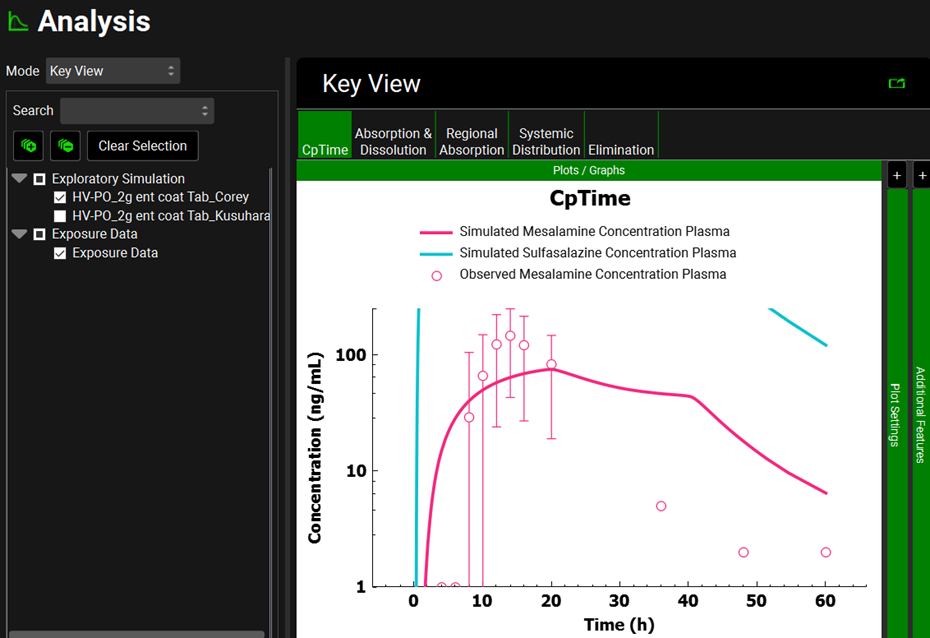
This plot has been formatted (it is colored by compound and the legend has been added). When following the instructions, the plot will be in the default formatting, that is, both lines and points are blue, and the legend is not displayed.
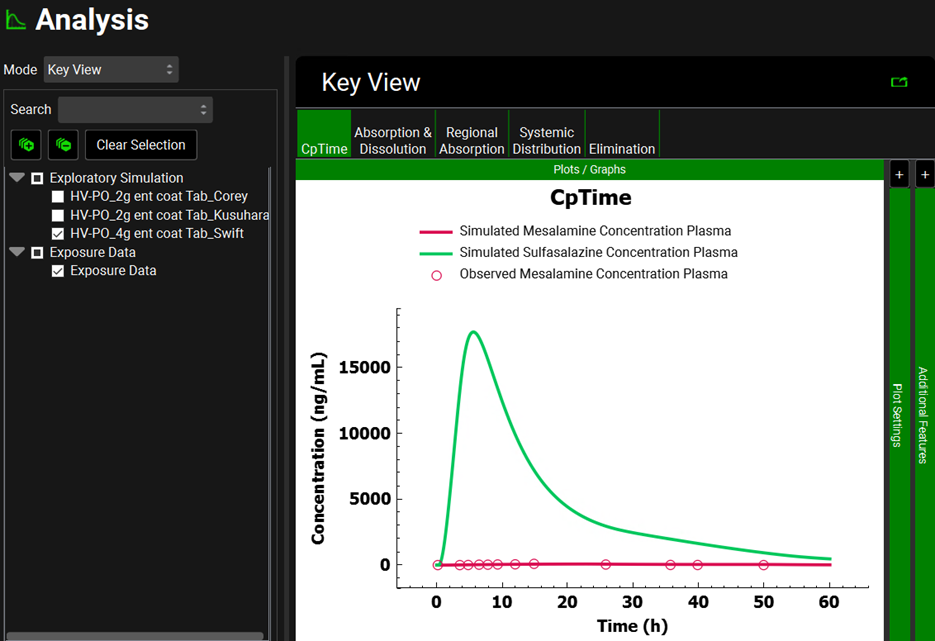
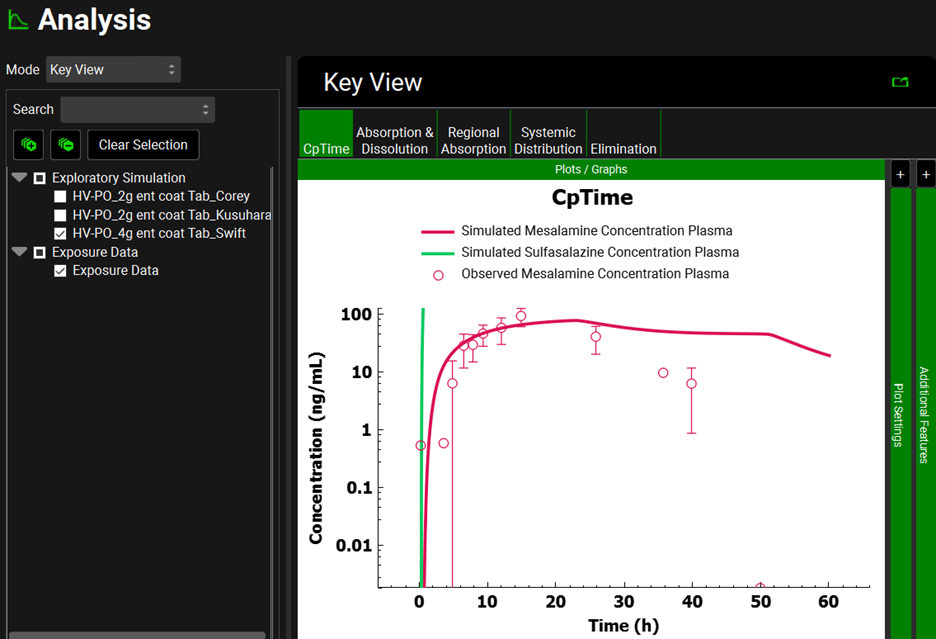
Repeat step 14 for Simulation “HV-PO_4g ent coat Tab_Swift”. Once in the Analysis view, you will see the simulated CpTime curve for mesalamine and sulfasalazine for a single dose of 4000 mg of sulfasalazine administered to healthy subjects as an enteric-coated tablet. The second screenshot represents the CpTime curves on a semi-logarithmic scale.
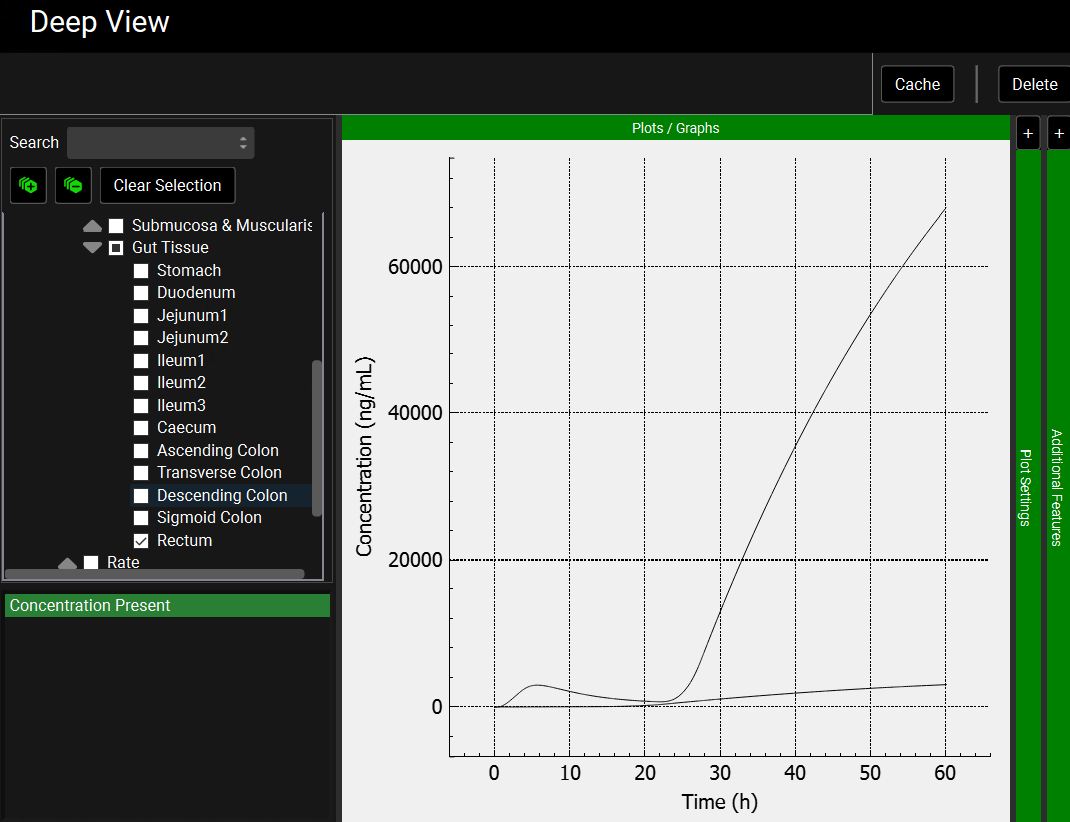
To view the simulated data for the gut tissues, switch the Mode to “Deep View” in the Analysis view. Select for example the concentration of Sulfasalazine in the rectum: Sulfasalazine_with_met > Gastrointestinal Tract > Concentration > Gut Tissue > Rectum from the tree and Concentration Present from the list. You can also select it for mesalamine: Mesalamine > Gastrointestinal Tract > Concentration > Gut Tissue > Rectum from the tree. You can observe the much lower concentration of mesalamine compared to sulfasalazine.