Tutorial: PKPlus™ Module
PKPlus™ is an optional module that quickly and easily fits parameters for one- to three-compartment PK models from single or multiple intravenous (iv) and/or oral plasma concentration-time data. Both linear and nonlinear simulations are supported.
In this tutorial we will cover the following topics:
PK models could also be fitted to iv data using the full ACAT model, just as fitting model to oral doses. The full ACAT model allows you to fit multiple data sets with complex kinetics at the same time, and has a more powerful nonlinear modeling capability; however, for simple data sets or rough estimates, PKPlus™ is much faster and fits all three models (1-, 2-, and 3-compartment) at once for quick comparisons.
This tutorial is displayed in "Light mode" to showcase the difference in themes.
Fit Compartmental PK using Single iv Cp-Time dataset
Open GPX™ and, in the Dashboard view, click on the icon next to Select to open an Existing project.
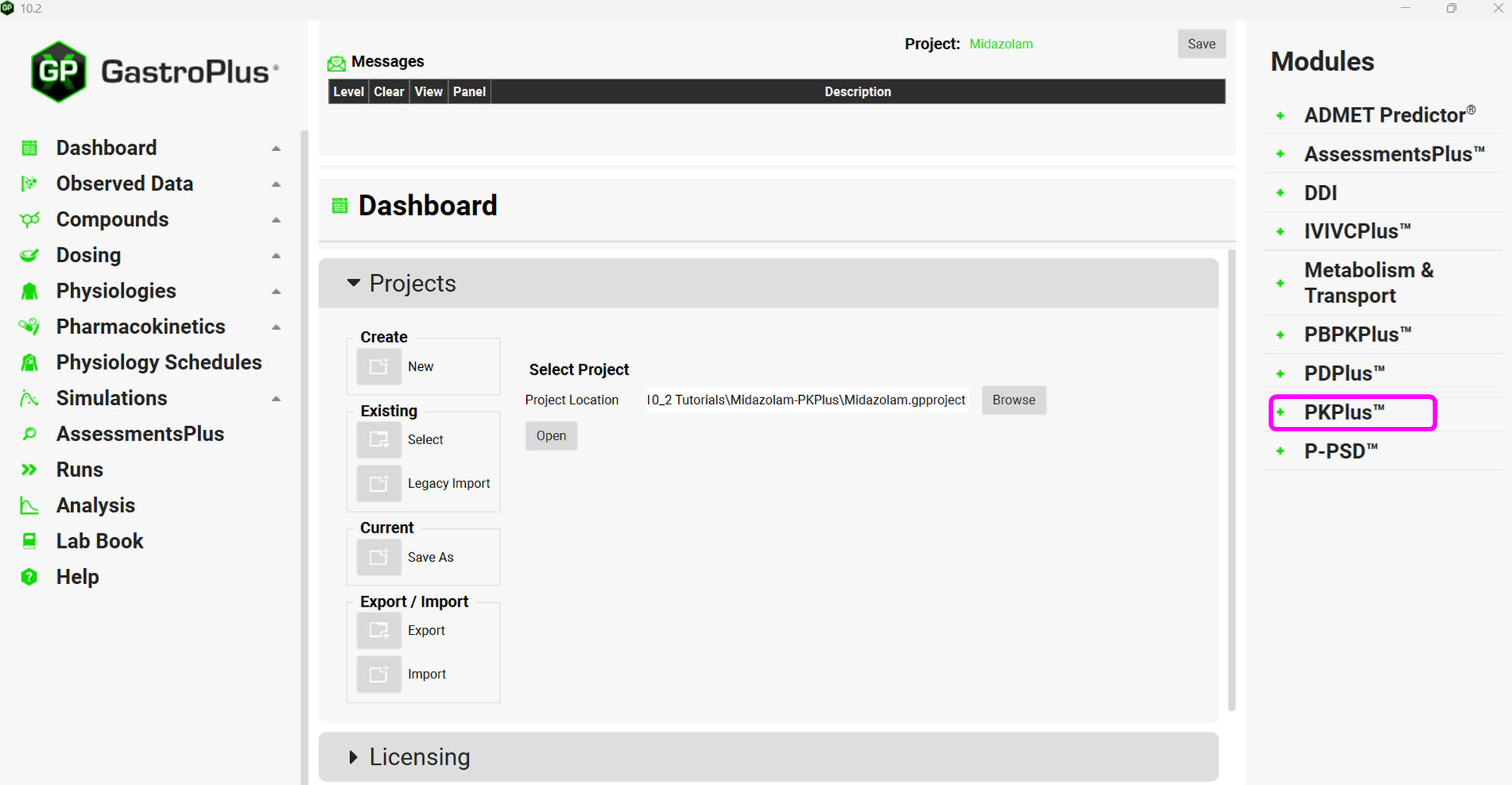
Click Browse and navigate to the C:\Users\<user>\AppData\Local\Simulations Plus, Inc\GastroPlus\10.2\Tutorials\Midazolam-PKPlus folder and select the project Midazolam.gpproject by clicking on it and clicking open.
Click on the Observed Data view in the navigation pane and then review the Exposure Data. This project contains Plasma Concentration versus Time profiles for iv and oral studies.
Click on PKPlus™ in the Modules pane located on the right-hand side of the interface.
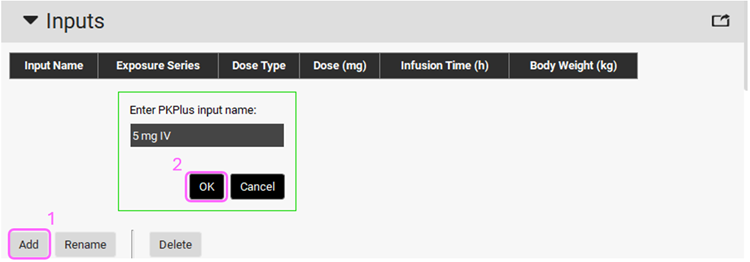
Now we need to set up the inputs for PKPlus™. Expand the Inputs panel by double clicking header or by clicking on the arrow. Click on Add button under the Inputs Table and name it “5 mg IV” and click OK (or press Enter).
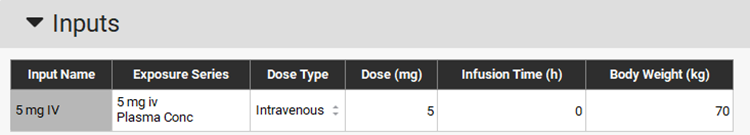
Select the Observed Data 5 mg iv as the Exposure Series and set Dose Type to Intravenous. The Dose and Body Weight are automatically obtained from the Observed Data. These inputs can also be manually modified.
Expand the Runs panel by double clicking header or by clicking on the arrow. Then create a run by clicking on Add button and name it “Single IV data run” and click OK or press Enter.
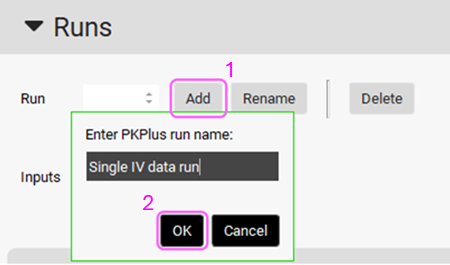
Select the 5 mg IV input from the Inputs frame by clicking on it (it will be highlighted in green).
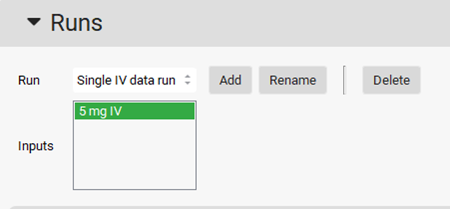
Under the Model Parameters sub-panel, select all the items in the Analyses section by clicking on each item.
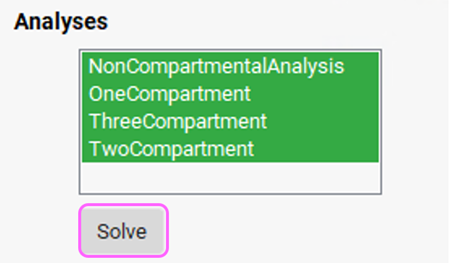
Click on the Solve button. PKPlus™ automatically fits noncompartmental, 1-, 2-, and 3-compartment pharmacokinetic models to the data and presents the fitted parameters for all models in the Results panel.
A message will appear in the Messages center informing the average body weight that was used to calculate volumes and clearances per kg. Click Done to Clear the message.
Once all selected models have been fitted, the best-fit model will be selected based on the lowest Akaike Information Criterion. The Best Fit toggle will be selected (green) to indicate the best fit.
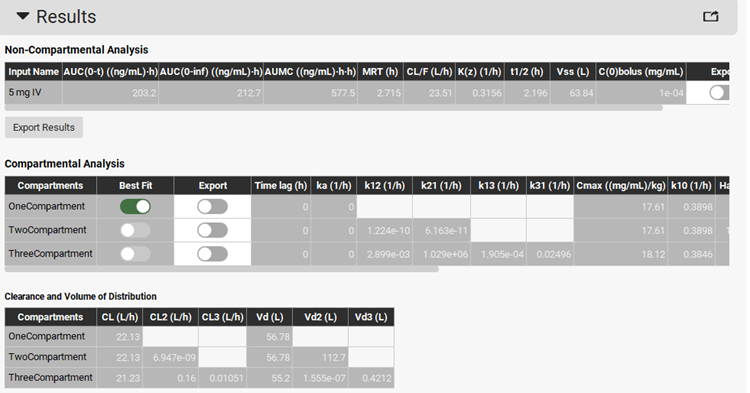
In this example, the one-compartment model has been selected as the best-fit model. Scrolling down, you can see the plot of the predicted PK profiles and Residuals for all three models. To compare the prediction of one-compartment model with the observed data, click on Observed 5 mg IV, then press Ctrl key and click on Predicted OneCompartment 5 mg IV.
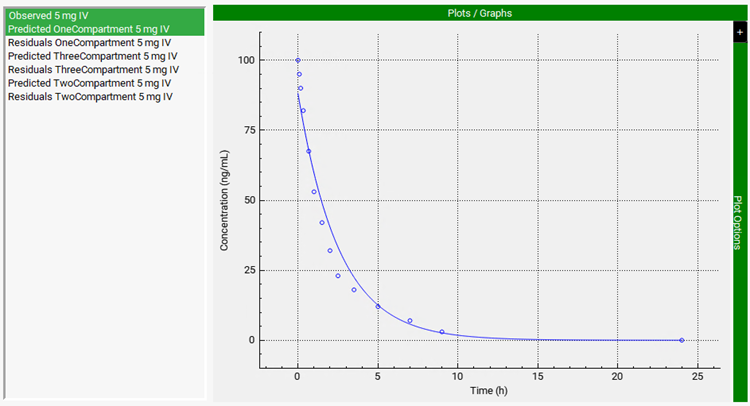
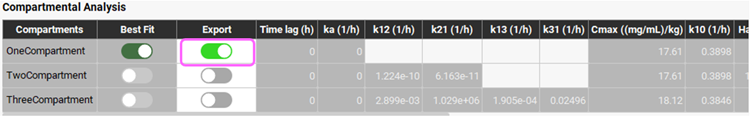
To export the fitted parameters of Best Fit model, select the Export toggle (it will be shown in green if it is on). Then scroll down past the Plots/Graphs and click on Export Results at the bottom of the view.
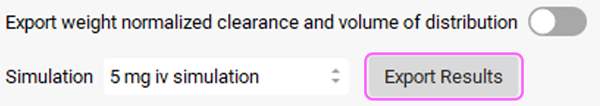
Since there is only one simulation in this project, 5 mg iv simulation will be automatically selected from the Simulation drop-down.
Two messages will appear in the Messages center informing that the Compartmental PK parameters have been exported to the Pharmacokinetics panel and that Simplified Absorption settings and first pass extraction have been exported to the selected simulation. Click Done on both messages to Clear them.
Navigate to the Pharmacokinetics view in the navigation pane. Expand the Compartmental panel if not already expanded and check the exported PK model named Single IV data run OneCompartment1 in the Compartmental panel.
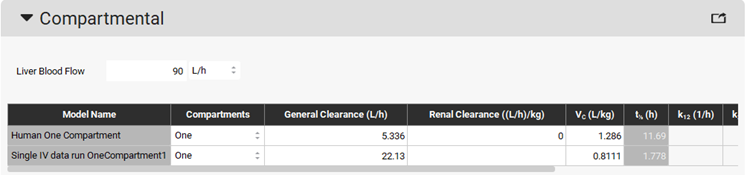
The Human One Compartment model (ADMET Predictor® module predicts the clearance and Vc for this model) can be deleted by pressing the Ctrl key and clicking on the first row, then clicking on the Delete button.
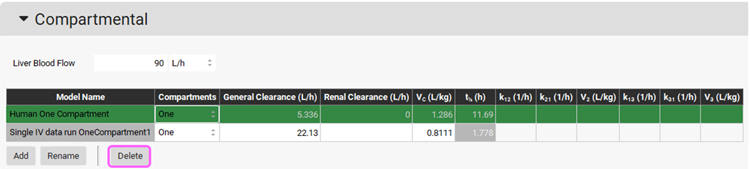
To check the simulation results of the exported compartment model, navigate to the Simulations view in the navigation pane, and expand the Compound Settings panel and make sure the Compartment Model Name has been set to Single IV data run OneCompartment1 for the 5 mg iv simulation.
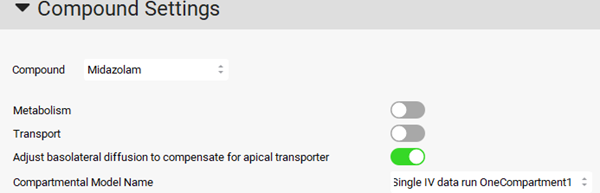
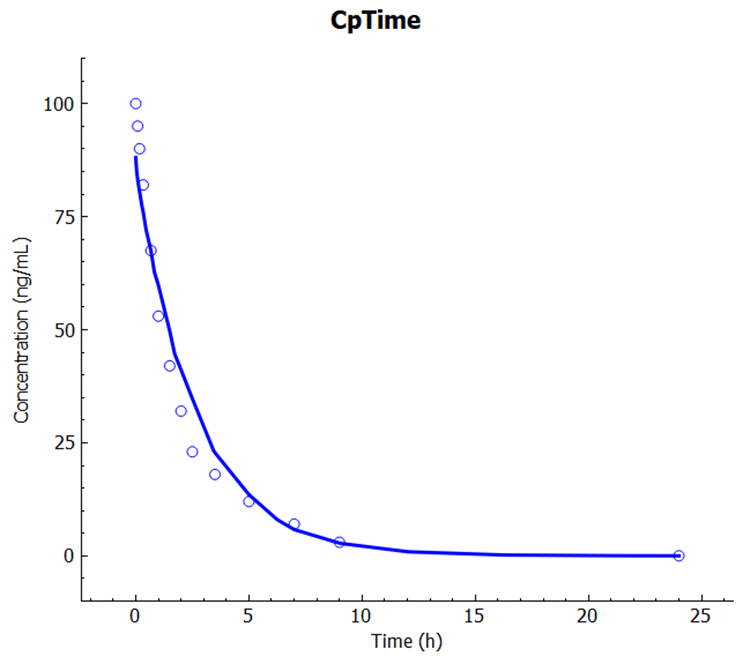
Click on Check Warnings then Run Simulation. Notice the good match between the predicted and observed plasma versus time profile in the Key View Cp-Time plot.
Fit Compartmental PK using multiple oral and iv Cp-Time datasets
Continue working with the Midazolam project. The observed data from the iv study (5 mg iv) and three oral studies (7.5 mg Solution, 15 mg Solution, and 30 mg Solution) will be used to fit the compartment model.
Click on PKPlus™ in the Modules pane located on the right-hand side of the interface.
Now we need to set up the inputs for these three oral studies in PKPlus™. Create another entry in the Inputs panel table by clicking on Add. Name the PKPlus™ input as 7.5 mg Solution. Select the 7.5mg Solution from the Group Name drop-down under Exposure Series and set the Dose Type to Oral.
Create entries for 15 mg Solution and 30 mg Solution linking each entry to their respective Exposure Series, as shown in the screenshot below.
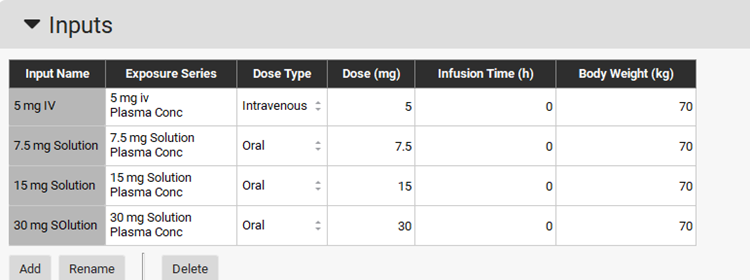
Scroll down to the Runs panel and create a new run named IV and Oral run, which will incorporate all 4 inputs for the fitting. Press Ctrl key while selecting multiple inputs.
Expand the Model Parameters sub-panel to see that there are three options for fitting the data: Use Individual Body Weights, Fit F% For Oral Doses, and Fit Lag Time For Oral Doses. Leave them off for the time being. We will return to these options later to examine their effect on the results of fitting.
Under the Model Parameters panel, select all the items in the Analyses section by clicking on each item and then click on Solve.
PKPlus™ automatically fits noncompartmental, 1-, 2-, and 3-compartment pharmacokinetic models to the four datasets and presents the fitted parameters for all models in the Results panel.
Note that the Two Compartmental model has been selected as the Best Fit model.
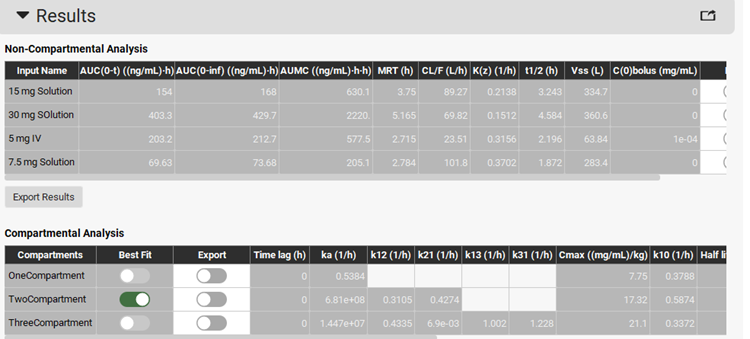
Scrolling down, you can see the plot of the predicted PK profiles and Residuals for all three models. To compare the prediction of Two-Compartment model with the observed data, press Ctrl key and click on all the observed data and the Predicted results for the Two Compartment model.
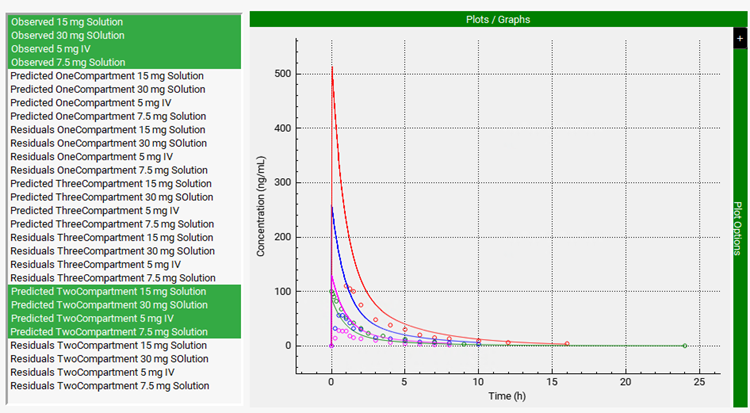
Notice that there is not a good fit between observed and simulated profiles (especially for the oral doses). This is a result of fitting options we have selected for the optimization. We chose not to fit the bioavailability for oral doses (the toggle of Fit F% for Oral Doses is turned off). Midazolam has a significant first pass extraction due to CYP3A4 mediated metabolism in gut and liver, which needs to be taken into consideration when fitting pharmacokinetics to oral doses.
Return to the Model Parameters sub-panel and select the Fit F% For Oral Doses toggle (it will be shown green if it is on).

Click on the Solve button to rerun the optimization.
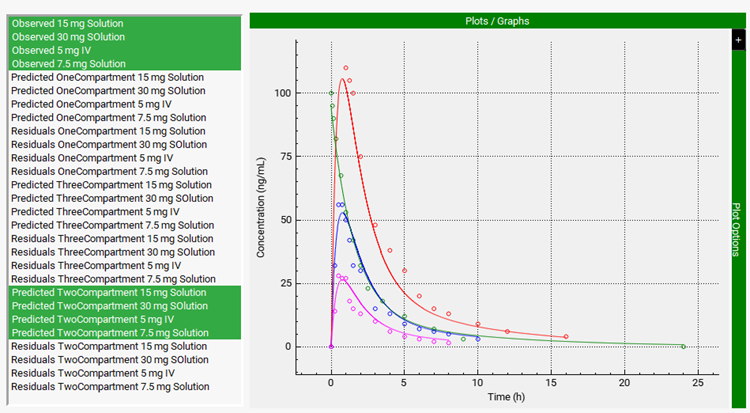
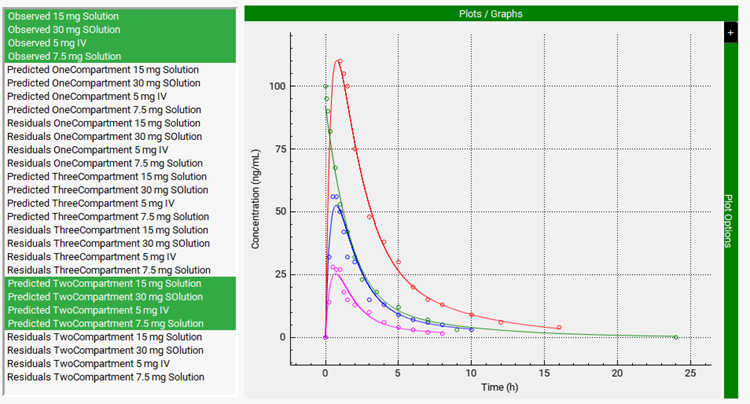
A two-compartment model was again selected as the best-fit model based on Akaike Information Criterion. Scroll down to the Graph and select all the Observed data and the simulated results for the Two-Compartment model. This time the agreement between simulated and observed profiles is much better.
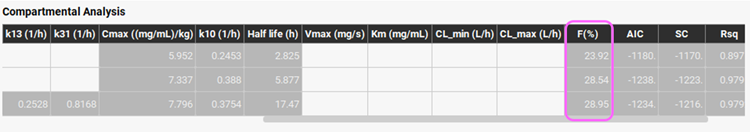
Examine the optimization results in the Compartmental Analysis table. Slide the bar at the bottom of the Compartmental Analysis table to see the fitted bioavailability, which is only ~28%. This is in excellent agreement with the literature value for a 15 mg oral dose of Midazolam.
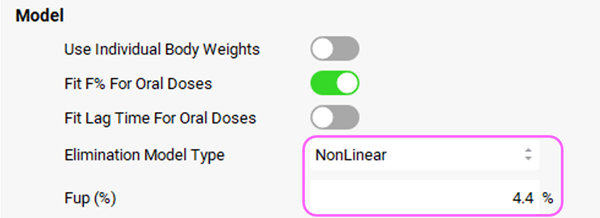
PKPlus™ also allows fitting of nonlinear kinetics. Scroll back up to the Model Parameters sub-panel and select NonLinear from the Elimination Model Type drop-down. Set the Fup (%) to 4.4%. To fit the correct intrinsic clearance, it is important to provide the correct unbound concentration, especially for highly bound drugs.
Click on the Solve button to rerun the optimization.
A Two-Compartment model was again selected as the Best Fit model based on Akaike Information Criterion. Scroll down to the Graph and select all the Observed data and the simulated results for the Two-Compartment model. The simulated results and the observed data have a good match.
Examine the optimization results in the Compartmental Analysis table. Slide the bar at the bottom of the Compartmental Analysis table to see the fitted bioavailability, which is ~28%.
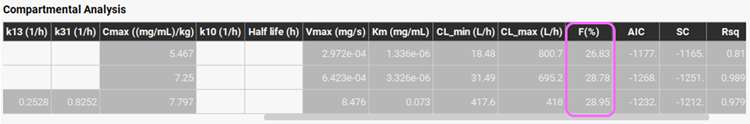
The fitted intrinsic clearance (CL_max (L/h)) for the Two-Compartment model is Vmax/Km = 695 L/h. This is in pretty good agreement with the intrinsic clearance calculated from in vitro Km and Vmax converted to in vivo values which is 943 L/h.
