Runs View
The Runs view is comprised of the Runs table, Simulation Settings, Output, and Run Controls panels below. See
Runs Table
The Runs table allows control over the types of runs to be conducted, as well as the simulation(s) to be used by each run. Various run types can be added: Simulations, Parameter Sensitivity Analysis (PSA), Population Simulations, Optimization, or Virtual Bioequivalence (VBE). The simulations to include in a given run can be selected using the toggles on the right side of the table.
Runs view, Runs table
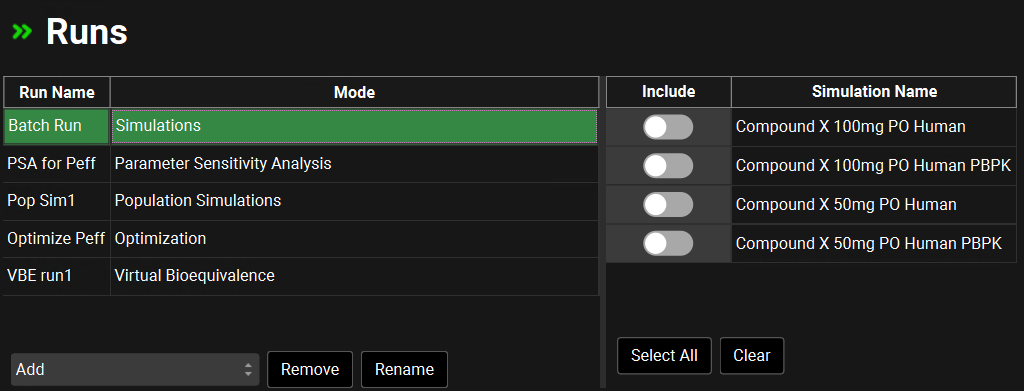
Input/Option | Description |
Run Name | The name of the run. Name is set when the run is created. |
Mode | Indicates the mode of run: Simulations (a batch of single simulations), Parameter Sensitivity Analysis (PSA), Population Simulations, Optimization, or Virtual Bioequivalence (VBE). The run mode is set when the run is created and cannot be changed. |
Add | Adds a new run. Clicking this will open a drop-down, from which the desired run mode is selected. |
Remove | Deletes the run currently selected in the table above. |
Rename | Renames the run currently selected in the table above. |
Include | A toggle. Controls whether the simulation (to the right) will be included in the run selected on the left. If the toggle is on (green) the simulation will be included in the run. If the toggle is off (grey) the simulation will not be included in the run. |
Simulation Name | The name of the Simulation created in the Simulations view. |
Select All | Sets all toggles to on, including all simulations in the project in the run. |
Clear | Sets all toggles to off. |
Simulation Settings panel
The Simulation Settings panel provides control over additional settings specific to PSA, Population Simulation, Optimization, and VBE. While there is significant overlap between the settings for these run modes, there are also different settings available for each. The unique Simulation Settings panels for each run mode are described individually below.
This panel is disabled for Simulations runs, which are a batch of single simulations, and do not require additional settings.
All run settings, including the selection of simulations, the parameters for PSA, Population Simulation, Optimization, and VBE run modes, and the output settings, are saved with the project and will be available in future sessions.
Simulation Settings panel for PSA
For parameter sensitivity analysis runs, the simulation settings panel provides control over the PSA type, parameter synchronization preferences, and selection and options for parameter permutation.
Runs view, Simulation Settings panel (for PSA run mode)
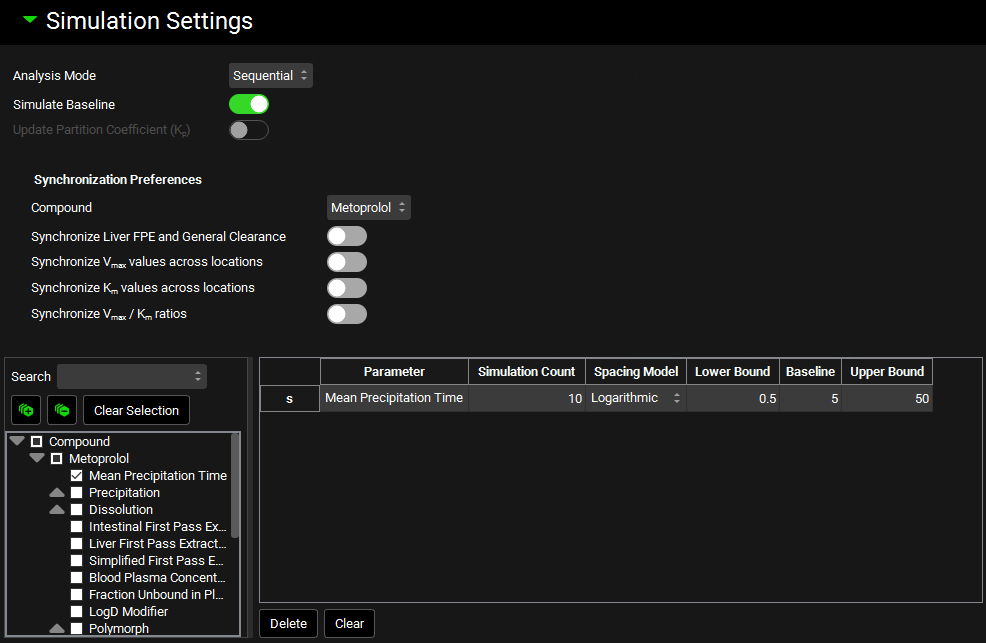
Input/Option | Description |
Analysis Mode | Selects the analysis mode for the PSA: sequential or cross. Sequential PSAs vary one parameter at a time, whilst keeping the other parameters at their baseline values (if more than one parameter is selected). This is equivalent to running a PSA on each parameter individually. Cross PSAs vary parameters in a pairwise fashion. |
Simulate Baseline | A toggle. Controls whether a baseline simulation will be run in addition to the simulations with permuted parameters. |
Update Partition Coefficient | A toggle. Controls whether tissue:plasma partition coefficients (Kps) will be updated during the PSA. This is relevant if parameters used in the calculation of Kps (e.g. pKa, LogD, Fup, Rbp) are varied as part of the PSA. This option is enabled only for PSAs using PBPK simulations. |
Synchronization Preferences | Controls synchronization of clearance and/or kinetic parameters when one or more related parameters are varied as part of the PSA. These settings allow related or interdependent parameters to be varied in equal amounts. |
Compound | Selects the compound to which the synchronization parameters will be applied. |
Synchronize Liver FPE and General Clearance | A toggle. If toggled on, the PSA run will re-estimate fixed Liver First Pass Extraction from General Clearance when general clearance is varied. Only applies to compartmental models using General Clearance. Both General Clearance and Liver FPE must be selected as parameters below. |
Synchronize Vmax values across locations | A toggle. If toggled on, the PSA run will synchronize Vmax scaling factors for the selected enzyme/transporter across various locations, ensuring they are each varied by the same amount. Only applies to Vmax scaling factors in locations selected for permutation. If there are multiple enzymes/transporters for the compound, and the Vmax scaling factors are selected in multiple locations for multiple enzymes/transporters, each enzyme/transporter is synced separately (across all locations where it is expressed). |
Synchronize Km values across locations | A toggle. If toggled on, the PSA run will synchronize Km scaling factors for the selected enzyme/transporter across various locations, ensuring they are each varied by the same amount. Only applies to Km scaling factors in locations selected for permutation. If there are multiple enzymes/transporters for the compound, and the Km scaling factors are selected in multiple locations for multiple enzymes/transporters, each enzyme/transporter is synced separately (across all locations where it is expressed). |
Synchronize Vmax/Km ratios | A toggle. If toggled on, the PSA run will synchronize the Vmax/Km scaling factor ratio for each metabolism/transport. The goal of this option is to maintain the intrinsic clearance of the kinetic process while changing the saturation point. Only applies if both the Vmax and the Km scaling factors are selected for permutation. |
Search | A text entry box. Used to search the parameter selection tree for parameters by name. To reset the parameter tree, clear the text in the search box and press Enter. |
Parameter Selection Tree | A hierarchy tree to select parameters for variation. Individual parameters can be identified by expanding and browsing the tree, or by using the search bar at the top. Parameters are selected/deselected using the check boxes. Once selected, parameters will appear in the table on the right. The available parameters are determined based on the inputs to the simulation(s) selected for the run. This tree selector functions the same way in PSA, Population Simulation, Optimization, and VBE Run modes. |
[+] and [-] buttons | Expand or collapse the entire parameter selection tree. |
Clear Selection | Unchecks the checkboxes for all parameters. Will also clear the table to the right. |
Parameter | The name of the parameter(s) being varied in the PSA run. |
Simulation Count | The number of simulations to be run for each parameter. The value can be edited. The default value is 10. |
Spacing Model | Selects whether the spacing of the parameter values will be uniform or logarithmic. |
Lower Bound | The lower bound of the parameter range to be tested. The first value will equal this lower bound, with subsequent values being based on the Spacing Model and Upper Bound. The initial lower bound is calculated relative to the baseline value specified in the simulation. This value can be edited. Note: If the baseline value is manually edited in the PSA table, the bounds do not get automatically updated. |
Baseline | The baseline value of the parameter, based on the simulation input. This value can be edited. Editing this value will only have an effect if Simulate Baseline is turned on. Changes to this value apply within this PSA run only. Note: The bounds must be edited separately. |
Upper Bound | The upper bound of the parameter range to be tested. The final value will equal this upper bound, with previous values being based on the Spacing Model and Lower Bound. The initial upper bound is calculated relative to the baseline value specified in the simulation. This value can be edited. Note: If the baseline value is manually edited in the PSA table, the bounds do not get automatically updated. |
Delete | Deletes the selected parameter from the table and deselects the parameter in the parameter selection tree on the left. Hold Ctrl-key and click to select the parameter (the row will be highlighted in green). |
Clear | Deletes all parameters from the table and deselect all parameters in the parameter selection tree on the left. Click Clear then click anywhere in the table to complete the action. |
If the property to be varied is calculated, as is the case for the Solubility Based Bile Salt Solubilization Ratio, the input needs to be switched to User Defined to enable it to be selected.
When multiple Forms are present in the selected simulations and the Forms fractions are selected to be permutated, the iteration file will list the values generated by the run mode. These fractions assigned by the run are then normalized in the engine when the simulation is executed. The normalized values can be seen by outputting the Inputs.
Simulation Settings panel for Population Simulations
For Population Simulation Runs, the Simulation Settings panel provides control over the sampling of the population, parameter synchronization preferences, and selection and options for individual parameter sampling.
Only one simulation can be selected as the baseline simulation for a Population Simulation Run.
A best practice is to name your Population Simulation Run name as succinctly (yet descriptive) as possible. There is a standard text limit on BE statistics excel files, hence, if the name is too long, the file won’t be generated.
The initial settings address the demographic sampling used to generate the virtual population. Additional PEAR sampling options are available for population simulation runs based on PBPK simulations. Synchronization of certain clearance and kinetic parameters is also available and behaves similarly to other run modes in which it is available. Parameter selection also behaves similarly but has sampling options unique to population simulations.
Runs view, Simulation Settings panel (Population Simulation run mode: General Settings)
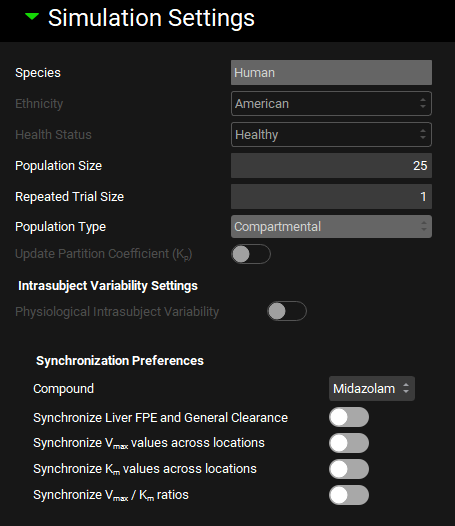
Input/Option | Description |
Species | The species of which the virtual population will be comprised. This is based on the simulation selected for the run and cannot be edited here. |
Ethnicity | A drop-down. Selects the ethnicity to be sampled in the population: American, Chinese, Japanese. Affects sampling of demographic information, as well as additional PEAR parameters for PBPK simulations. Initial value is based on the physiology schedule in the baseline simulation. The selection can be edited. |
Health Status | A drop-down. Selects the health status of the sampled population, Cirrhosis A, B, C, Healthy, Morbidly Obese, NonAlcoholic Fatty Liver Disease, NonAlcoholic Steato Hepatitis, Obese, Pregnant, Renal Impair End Stage, Renal Impair Mild, Renal Impair Moderate, Renal Impair Severe. Affects PEAR parameters for PBPK simulations. Initial value is based on the physiology schedule in the baseline simulation. The selection can be edited. |
Population Size | The number of subjects to include in the virtual population. The value can be edited. Note: If a population is selected in the Generate Populations Settings table below, this selection will be disabled. |
Repeated Trial Size | The number of repeat trials to be conducted. Each additional trial will resample a new population within the same parameter ranges. The value can be edited. Note: If a population is selected in the Generate Populations Settings table below, this selection will be disabled. |
Population Type | A drop-down. Selects which population sampling method to use. If the baseline simulation uses a compartmental PK model, only compartmental populations will be available. If the baseline simulation is a PBPK simulation, the options are PEAR or Monte Carlo, with PEAR being the default. |
Update Partition Coefficient | A toggle. Controls whether tissue:plasma partition coefficients (Kps) will be updated during the Population Simulation. This is relevant if parameters used in the calculation of Kps (e.g. pKa, LogD, Fup, Rbp) are varied. This option is enabled only for simulations using a PBPK model. Note: If this toggle is ON and the Kps are also selected for sampling, the sampled values will get overwritten by the calculated Kp. This value can be seen by outputting the Inputs. |
Intrasubject Variability Settings | Section where intrasubject variability is set up. Applicable only when there are changes in physiology in the physiology schedule for the selected simulation.
|
Synchronization Preferences | Controls synchronization of clearance and/or kinetic parameters when one or more related parameters are sampled as part of the population simulation. These settings allow related or interdependent parameters to be sampled equally. |
Compound | A drop-down. Selects the compound to which the synchronization parameters will be applied. |
Synchronize Liver FPE and General Clearance | A toggle. If toggled on, the population simulation run will re-estimate fixed Liver First Pass Extraction from General Clearance when general clearance is sampled. Only applies to compartmental models using General Clearance. Both General Clearance and Liver FPE must be selected as parameters below. |
Synchronize Vmax values across locations | A toggle. If toggled on, the population simulation run will synchronize Vmax scaling factors for the selected enzyme/transporter across various locations, ensuring they are each changed by the same amount. Only applies to Vmax scaling factors in locations selected for sampling. |
Synchronize Km values across locations | A toggle. If toggled on, the population simulation run will synchronize Km scaling factors for the selected enzyme/transporter across various locations, ensuring they are each changed by the same amount. Only applies to Km scaling factors in locations selected for sampling. |
Synchronize Vmax/Km ratios | A toggle. If toggled on, the population simulation run will synchronize the Vmax/Km scaling factor ratio for each metabolism/transport. The goal of this option is to maintain the intrinsic clearance of the kinetic process while changing the saturation point. Only applies if both the Vmax and the Km scaling factor are selected for sampling. |

Note: for synchronize parameters to function correctly, all parameters to synchronize must be selected, see the Population Simulation Parameter Selection.
When multiple Forms are present in the selected simulations and the Forms fractions are selected to be permutated, the iteration file will list the values generated by the run mode. These fractions assigned by the run are then normalized in the engine when the simulation is executed. The normalized values can be seen by outputting the Inputs.
Human PEAR Settings Sub-panel
Once the initial settings have been set, additional options are available in the Human PEAR Settings sub-panel, where users can specify the percentage of the population that is male and the PEAR physiology sampling ranges for Age, Weight, Height, Body Mass Index (BMI), and Enzyme Phenotype Frequency. These settings are only available for human PBPK population simulations.
Runs view, Simulation Settings panel (Population Simulation run mode), Human PEAR Settings sub-panel
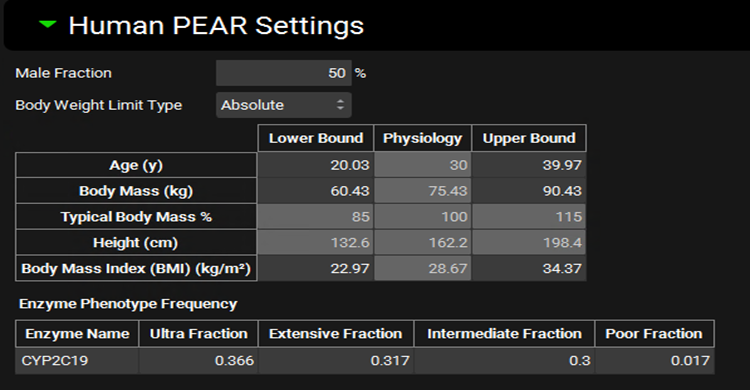
Input/Option | Description |
Male Fraction | The proportion of subjects, as a percentage, of the virtual population that will be male. The remainder will be female. The value can be edited. |
Body Weight Limit Type | A drop-down. Selects whether sampling of body mass will be bounded by absolute numbers or by percentage of baseline body mass. This will control which of those range options is enabled below. |
Age | The range for sampling the age, in years, of subjects in the virtual population. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is provided for informational purposes only. |
Body Mass | The range for sampling body mass, in kg, of subjects in the virtual population. This option is enabled only if Body Weight Limit Type is set to Absolute. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is provided for informational purposes only. |
Typical Body Mass Percent | The range for sampling body mass, as a percentage of baseline, of subjects in the virtual population. This option is enabled only if Body Weight Limit Type is set to Percentage. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is used to determine the numerical bounds based on the percentages entered and is provided for information only. |
Height | The range for sampling the height, in cm, of subjects in the virtual population. This value is calculated from body mass and BMI and changes in either body mass or BMI will cause Height to be automatically recalculated. It cannot be edited and is provided for information only. |
Body Mass Index (BMI) | The range for sampling the BMI, in kg/m2, of subjects in the virtual population. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is provided for informational purposes only. |
Enzyme Phenotype Frequency | This section controls the relative frequency of phenotype occurrence for relevant enzymes in the virtual population. The actual expression of each enzyme will still be sampled (if selected as a parameter for sampling below), but the baseline value from which the sampling is done will be based on the phenotype. The chance of a given subject having a given phenotype is set here. This section will only be active for enzymes active in the baseline simulation which have kinetics defined for two or more phenotypes in the enzyme kinetics table. |
Enzyme Name | The name of the enzyme for which the phenotype frequencies will apply. |
Ultra, Extensive, Intermediate, and Poor Fractions | The relative frequencies, as fractions, of the respective enzyme phenotypes. The default values are based on available literature information for enzyme phenotype distributions in the given ethnicity. These values can be edited but must add up to one. |
Runs view, Simulation Settings panel (Population Simulation run mode), Human PEAR Settings sub-panel for pregnant physiologies
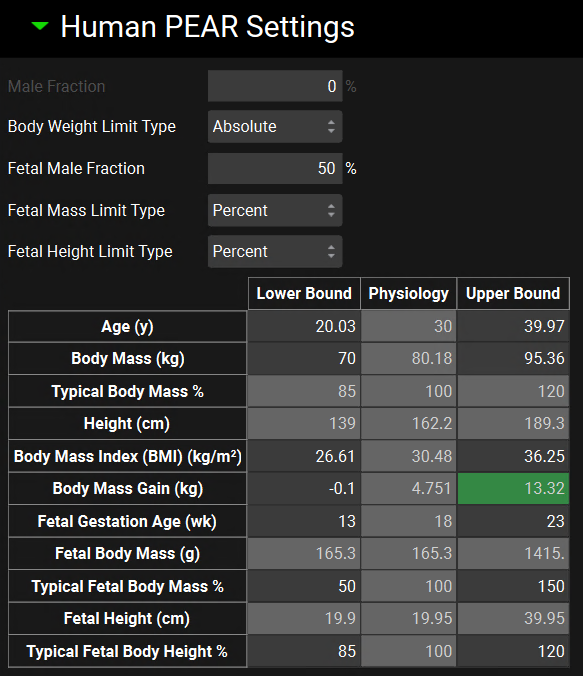
Input/Option | Description |
Male Fraction | Disable since the pregnant physiology is always Female. Value is set to zero which means 100% of the individuals will be female. |
Body Weight Limit Type | A drop-down. Selects whether sampling of body mass will be bounded by absolute numbers or by percentage of baseline body mass. This will control which of those range options is enabled below. |
Fetal Male Fraction | The proportion of subjects, as a percentage, of the virtual population that will be male. The remainder will be female. The value can be edited. |
Fetal Mass Limit Type | Displays the sampling of fetal body mass which will be bounded by percentage of baseline body mass. |
Fetal Height Limit Type | Displays the sampling of fetal height which will be bounded by percentage of baseline body mass. |
Age | The range for sampling the age, in years, of subjects in the virtual population. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is provided for informational purposes only. |
Body Mass | The range for sampling body mass, in kg, of subjects in the virtual population. This option is enabled only if Body Weight Limit Type is set to Absolute. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is provided for informational purposes only. |
Typical Body Mass Percent | The range for sampling body mass, as a percentage of baseline, of subjects in the virtual population. This option is enabled only if Body Weight Limit Type is set to Percentage. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is used to determine the numerical bounds based on the percentages entered and is provided for information only. |
Height | The range for sampling the height, in cm, of subjects in the virtual population. This value is calculated from body mass and BMI and changes in either body mass or BMI will cause Height to be automatically recalculated. It cannot be edited and is provided for information only. |
Body Mass Index (BMI) | The range for sampling the BMI, in kg/m2, of subjects in the virtual population. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is provided for informational purposes only. |
Body Mass Gain | The range for sampling the body mass gain, in kg, of subjects in the virtual population. This value is calculated from the baseline physiology in the simulation. It cannot be edited and is provided for information only. |
Fetal Gestation Age | The range for sampling fetal gestation age, in weeks. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is used to determine the numerical bounds and is provided for information only. |
Fetal Body Mass | The range for sampling body mass, in g, of subjects in the virtual population. This option is not editable. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is provided for informational purposes only. |
Typical Fetal Body Mass % | The range for sampling body mass, as a percentage of baseline, of subjects in the virtual population. This option is the default and is always enabled. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is used to determine the numerical bounds based on the percentages entered and is provided for information only. |
Fetal Height | The range for sampling the height, in cm, of subjects in the virtual population. It cannot be edited and is provided for information only. |
Typical Fetal Body Height | The range for sampling height, as a percentage of baseline, of subjects in the virtual population. This option is the default and is always enabled. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is used to determine the numerical bounds based on the percentages entered and is provided for information only. |
Population Simulation Parameter Selection
The parameter selection tree and table behave similarly to the PSA and Optimization run modes. However, for population simulations, a list of parameters is selected by default, based on the parameters expected to vary in a population. The baseline values for these parameters are derived from the baseline simulation for the population simulation run. The coefficient of variation (CV) values are based on literature data where available, with the default being 10% for the remainder. This CV is applied to the baseline value from the baseline simulation using the indicated distribution type to sample the parameter value that will be applied. A new sampling is done for each subject to achieve the desired distribution.
Of note, for PEAR populations, physiological parameters calculated based on the PEAR physiology will have the “PEAR” distribution type available and selected by default. This method first recalculates the baseline value based on the sampled subject’s PEAR physiology (see Human PEAR Sampling above). Sampling is then conducted for each subject using their unique baseline value and a log-normal distribution type, utilizing the indicated CV. This ensures that sampling of physiological parameters aligns with the subject’s demographic information.
Runs view, Simulation Settings panel (Population Simulation: Parameter Selection)
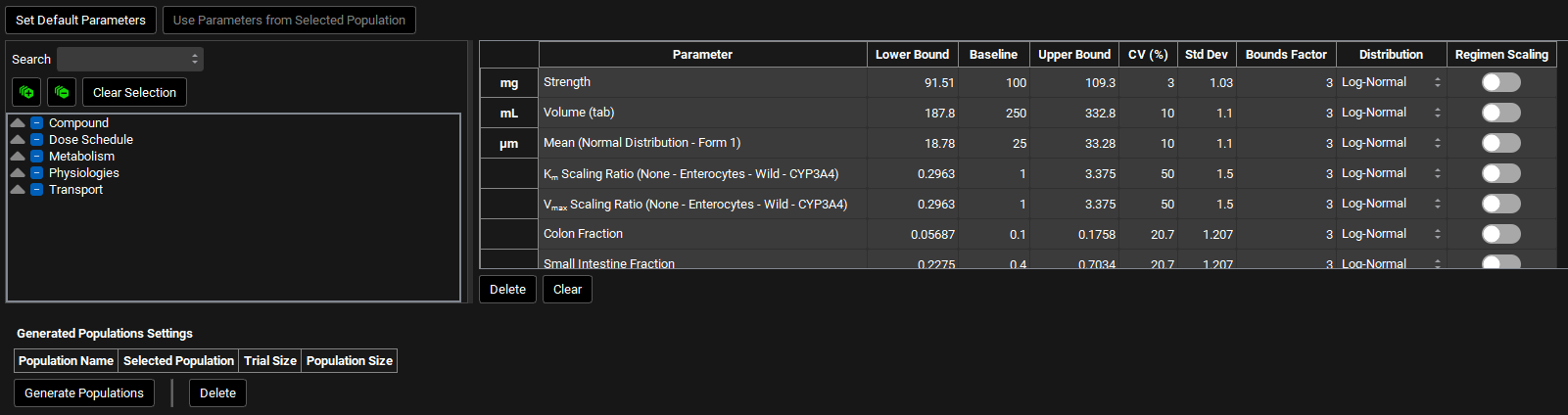
Input/Option | Description |
Set Default Parameters | Resets the parameter selection and values, CV %, Bounds Factor and Distribution to the GastroPlus® defaults. |
Use Parameters from Selected Population | Resets the parameters table to reflect the population selected in the Generated Populations Settings table below. |
Parameter Selection Tree | A hierarchy tree for selecting parameters for sampling. Individual parameters can be identified by expanding and browsing the tree, or by using the search bar at the top. Parameters are selected/deselected using the check boxes. Once selected, parameters will appear in the table on the right. The available parameters are determined based on the inputs to the simulation(s) selected for the run. A variety of relevant parameters are already selected by default for population simulation runs. This tree selector functions the same way in PSA, Population Simulation, Optimization, and VBE Run modes. |
Search | A text entry field. Used to search the parameter selection tree for parameters by name. To reset the parameter tree, clear the text in the search box and press Enter. |
[+] and [-] buttons | Expand or collapse the entire parameter selection tree. |
Clear Selection | Unchecks the checkboxes for all parameters and clears the table to the right. |
Parameter | The name of the parameter(s) being sampled in the population simulation run. |
Lower Bound | The lower bound of the sampling range, as determined by the baseline value, the CV, and the distribution type. This value is provided for informational purposes only; it cannot be edited. |
Baseline | The baseline value for the parameter, derived from the baseline simulation. This value can be edited to override the default baseline and the bounds will be automatically recalculated based on the baseline value if changed. Note that for parameters using the PEAR Sampling distribution type editing this value will have no effect as this value is recalculated internally for each subject based on their sampled demographic information. |
Upper Bound | The upper bound of the sampling range, as determined by the baseline value, the CV, and the distribution type. This value is provided for informational purposes only; it cannot be edited. |
CV (%) | The coefficient of variation, in percent, of the parameter distribution. Default values are based on literature information where available; other parameters have a default of 10%. This value can be edited. Editing this value will trigger a recalculation of the Std Dev and the Lower and Upper Bounds. This value will be automatically recalculated if Std Dev is edited. |
Std Dev | The standard deviation of the parameter distribution. The default value is calculated based on the default CV. This value can be edited. Editing this value will trigger a recalculation of the CV and the Lower and Upper Bounds. This value will be automatically recalculated if CV is edited. |
Bounds Factor | The number of standard deviations describing the sampling range. This value affects the Lower and Upper Bounds and can be edited. |
Distribution | Selects the distribution type for sampling. Options include Uniform, Normal, Log-Normal, and, if applicable, PEAR. The default selection for all non-PEAR parameters is Log-Normal. |
Regimen Scaling | A toggle that indicates gut physiological parameters (transit times, intestinal fluid volume, pH values, and bile salt concentrations for each gut section) that will be scaled in the second period. |
Delete | Deletes the selected parameter from the table and deselects the parameter in the parameter selection tree on the left. |
Clear | Deletes all parameters from the table and deselects all parameters in the parameter selection tree on the left. |
Population Name | Name given by the user for the generated population. |
Selected Population | A toggle. Indicates which population will be used in the run. |
Trial Size | The number of repeat trials to be conducted in the run. This value is set up in the “Repeated Trial Size” field above. |
Population Size | The number of subjects included in the virtual population. This value is set up in the “Population Size” field above. |
Generate Populations | A button that creates a population based on the previous selections. |
Delete | A button. Deletes the highlighted population. |
Simulation Settings panel for Optimization
For optimization runs, the simulation settings panel provides control over parameter synchronization preferences. Selection of parameters and data for optimization are handled in the Optimization Parameter Selection and Observation Selection and Weights sub-panels below.
Optimization runs can include multiple simulations, but only parameters common to all simulations can be optimized, and all simulations must use the same type of PK model.
See Optimization View for information about transferring optimized parameter values back to the project.
Runs view, Simulation Settings panel (Optimization run mode)
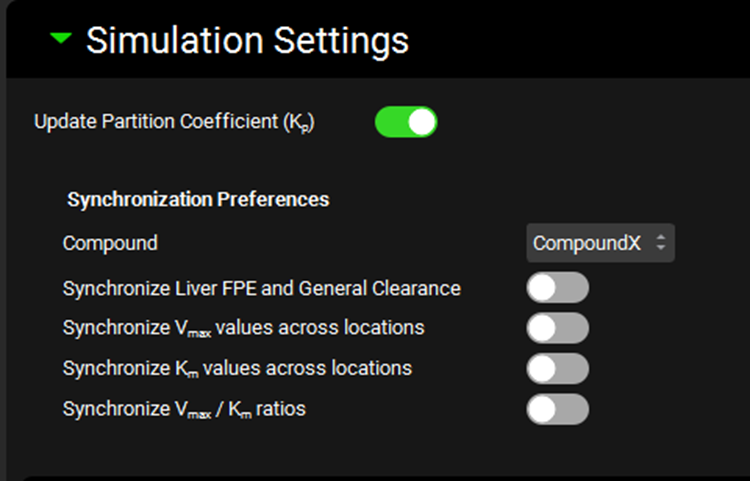
Input/Option | Description |
Update Partition Coefficient | A toggle. Controls whether tissue:plasma partition coefficients (Kps) will be updated during the optimization. This is relevant if parameters used in the calculation of Kps (e.g. pKa, LogD, Fup, Rbp) are being optimized. This option is enabled only for optimizations using a PBPK model. |
Synchronization Preferences | Controls synchronization of clearance and/or kinetic parameters when one or more related parameters are optimized. These settings allow related or interdependent parameters to be optimized in tandem. |
Compound | A drop-down. Selects the compound to which the synchronization parameters will be applied. |
Synchronize Liver FPE and General Clearance | A toggle. If toggled on, the optimizer will re-estimate fixed Liver First Pass Extraction from General Clearance as general clearance is optimized. Only applies to compartmental models using General Clearance. Both General Clearance and Liver FPE must be selected as parameters below. |
Synchronize Vmax values across locations | A toggle. If toggled on, the optimization run will synchronize Vmax scaling factors for the selected enzyme/transporter across various locations. Only applies to Vmax scaling factors in locations selected for optimization. If there are multiple enzymes/transporters for the compound, and the Vmax scaling factors are selected in multiple location for multiple enzymes/transporters, each enzyme/transporter is synced separately (across all locations where it is expressed). |
Synchronize Km values across locations | A toggle. If toggled on, the optimization run will synchronize Km scaling factors for the selected enzyme/transporter across various locations. Only applies to Km scaling factors in locations selected for optimization. If there are multiple enzymes/transporters for the compound, and the Km scaling factors are selected in multiple location for multiple enzymes/transporters, each enzyme/transporter is synced separately (across all locations where it is expressed). |
Synchronize Vmax/Km ratios | A toggle. If toggled on, the optimization run will synchronize the Vmax/Km scaling factor ratio for the selected metabolism/transport. The goal of this option is to maintain the intrinsic clearance of the kinetic process while changing the saturation point. Only applies if both the Vmax and the Km scaling factor are selected for optimization. |

Note: for synchronize parameters to function correctly, all parameters to synchronize must be selected, see the Optimization Parameter Selection sub-panel section.
Optimization Parameter Selection sub-panel
The Optimization Parameter Selection sub-panel permits selection of parameters to be optimized. Available parameters are based on the baseline simulation(s). Only parameters common to all baseline simulations should be selected for optimization when more than one simulation has been selected.
The parameter selection tree and table behave similarly to the PSA and Population Simulation run modes. No parameters are selected by default for optimization.
Runs view, Simulation Settings Panel (Optimization), Optimization Parameter Selection sub-panel
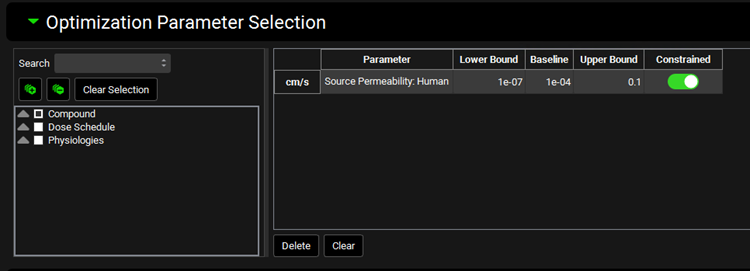
Input/Option | Description |
Parameter Selection Tree | A hierarchy tree for selecting parameters for optimization. Individual parameters can be identified by expanding and browsing the tree, or by using the search bar at the top. Parameters are selected/deselected using the check boxes. Once selected, parameters will appear in the table on the right. The available parameters are determined based on the inputs to the simulation(s) selected for the run. This tree selector functions the same way in PSA, Population Simulation, Optimization, and VBE run modes. |
Search | A text entry field. Used to search the parameter selection tree for parameters by name. To reset the parameter tree, clear the text in the search box and press Enter. |
[+] and [-] buttons | Expand or collapse the entire parameter selection tree. |
Clear Selection | Unchecks the checkboxes for all parameters. Will also clear the table to the right. |
Parameter | The name of the parameter(s) being optimized |
Lower Bound | The lower bound of search range. The initial value is based on the baseline value specified in the simulation. This value can be edited. Note: If the baseline value is manually edited in the Optimization table, the bounds do not get automatically updated. |
Baseline | The baseline value for the parameter, derived from the baseline simulation. This value can be edited to override the baseline value. Changes to this value apply within this Optimization run only. Note: The bounds must be edited separately. This value is used as the starting point for the optimization. |
Upper Bound | The upper bound of search range. The initial value is based on the baseline value specified in the simulation. This value can be edited. Note: If the baseline value is manually edited in the Optimization table, the bounds do not get automatically updated |
Constrained | A toggle. Controls whether the parameter will be constrained within the lower and upper bounds during the optimization. It is on by default. |
Delete | Deletes the selected parameter from the table. Will also deselect the parameter in the parameter selection tree on the left. |
Clear | Deletes all parameters from the table. Will deselect all parameters in the parameter selection tree on the left. Click Clear then click anywhere in the table to complete the action. |
If the property to optimize is calculated, as is the case for the solubility based bile salt solubilization ratio, the input needs to be switched to User Defined to enable it to be selected.
Observation Selection and Weights
The Observation Selection and Weights Sub-panel permits selection of observed data against which the optimization will be conducted. For each entry here, the corresponding simulated values will be calculated during the optimization process, with the goal of minimizing the error between the simulated and observed values.
There are two drop-downs for observed data. The top drop-down allows selection of observed data series (e.g. Cp-time profiles or tissue concentration-time profiles), while the bottom allows selection of summary pharmacokinetic parameters (e.g. Fa, Fdp, F, etc.). The series or summary parameters available are dependent on observed data linked in the baseline simulation(s). Once the Data Source has been selected in the respective drop-down, a list will appear below, and the parameter(s) (e.g. Venous Return, Liver or bioavailability (F)) can be selected. Selected parameters will be highlighted in green. The observed data selected for the optimization run will appear in the table to the right of the drop-downs, where the observation weight can be set.
For the observed data series, if both Systemic Circulation and Venous Return appear, the user can select both. Alternatively, for a simulation with a Compartmental pharmacokinetic model, the user should select only Systemic Circulation, or for a simulation with a PBPK pharmacokinetic model, the user should select only Venous Return.
Runs view, Simulation Settings Panel (Optimization), Observation Selection and Weights Sub-panel
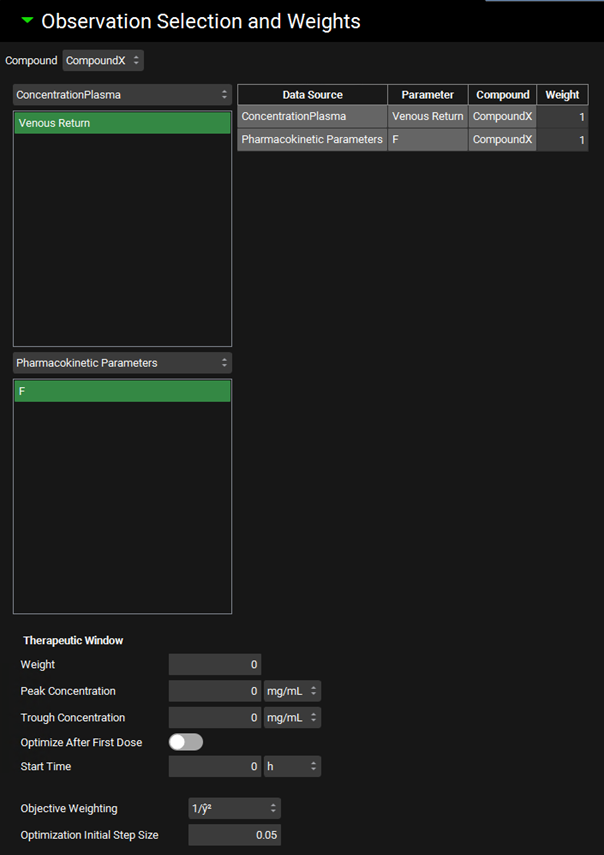
Input/Option | Description |
Compound | A drop-down. Selects the compound for which observed data can be selected for the optimization run. Observed data must first be linked to this compound in the baseline simulation(s). |
Data Source Drop-down: Series Type
| Lists the series available for selection. Typical options might include plasma or tissue concentration or mass present in urine. |
Parameter Selection: Series | Allows selection of specific series for use in optimization. Entries are selected by clicking, and multiple series may be used. Selected entries are highlighted in green and will appear in the table to the right. |
Data Source Drop-down: Parameters
| Lists the parameter entries available for selection. Currently only summary Pharmacokinetic Parameters are available. |
Parameter Selection: Parameters | Allows selection of specific summary parameters for use in optimization. Entries are selected by clicking, and multiple series may be used. Selected parameters are highlighted in green and will appear in the table to the right. |
Data Source | Indicates the data source type for the observation. Corresponds to the Series and Parameter Type Drop-downs. |
Parameter | Indicates the specific series or summary parameter. |
Compound | Indicates the compound to which the parameter applies. |
Weight | Gives the weight assigned to the observation, with a default value of 1. This value can be edited. The weight is used as a multiplier for the error between simulation and observed values for the parameter during calculation of the overall objective function during optimization. Increasing this value will increase the weight assigned to this error (and thus the impact on optimization), while decreasing this value will decrease the weight. A value of 0 will effectively ignore this observation during optimization. Observation weight can be used in two ways. First, it can be used to emphasize a given parameter, such as putting greater weight on plasma concentrations than mass in urine, if desired for a particular scenario. Second, it can be used to adjust for differences in units between observed data. For example, while concentrations are measured in mass per volume, bioavailability is measured as a percent, which can give undue weight to one scale if calculating the objective function using absolute error. As such, observation weight can be used to compensate for this phenomenon. |
Therapeutic Window | This section deals with target therapeutic plasma concentrations when optimizing dose-related parameters to a therapeutic range. |
Weight | The weight assigned to this measure. See Weight above. |
Peak Concentration | The target peak plasma concentration, in mg/mL. This value can be edited. |
Trough Concentration | The target trough plasma concentration, in mg/mL. This value can be edited. |
Optimize After First Dose | A toggle. Controls whether optimization to peak and trough concentrations will use the first peak and trough concentrations in the simulation (and every peak and trough thereafter), or if it will use peak and trough concentrations only after a specified time point. The former option is used if toggled on. |
Start Time | The time, in hours, after which peak, and trough concentrations will be used for therapeutic window optimization. The value can be edited. This option is useful for optimizing using only steady state concentrations. Applies only if Optimize After First Dose is toggled off. Therapeutic window optimization will only have a contribution if the simulation length exceeds this value. A Start Time of 0 will have the same effect as toggling Optimize After First Dose on. |
Objective Weighting | Selects the objective weighting function to be used in calculation of the objective function. Options are: Uniform (unity), 1/y, 1/y2, 1/ŷ, 1/ŷ2, 1/(y+ ŷ), 1/(y+ ŷ)2, 1/σ2 with σ from observed exposure data CV, 1/σ2 with σ calculated from observed exposure data, and 1/σ2 with σ interpolated from user specified CV. 1/ŷ2 is the default selection. |
Optimization Initial Step Size | The initial step size taken by the optimizer during the search function, as a fraction of the baseline value. The value can be edited. The default value is 0.05. For baseline values of 0, the first step will be a fixed value; subsequent steps will use this Optimization Initial Step Size value as the starting step size. |
Simulation Settings panel for Virtual Bioequivalence
For Virtual Bioequivalence (VBE) Runs, the Simulation Settings panel provides the option to select the study design (crossover vs parallel), control over the crossover design, sampling of the population, parameter synchronization preferences, and selection and options for individual parameter sampling. For GPX™ 10.2 there have been significant enhancements to the Virtual Bioequivalence runs mode. When opening projects created in GPX™ 10.1, please delete any VBE runs and recreate them to define all the new settings for all of the additional functionality.
For the non-replicate VBE study design, multiple simulations may be selected for VBE Runs, permitting the use of different baseline simulations for the reference and the test(s). The baseline simulation to use as the reference simulation is selected by the user (see below), while any other simulations included in the run will be considered as test simulations. All simulations must use the same type of PK model (Compartmental or PBPK).
The initial settings address the selection of the reference simulation, bioequivalence study design type and intrasubject variability.
A best practice is to name your Virtual Bioequivalence Run name as succinctly (yet descriptive) as possible. There is a standard text limit on BE statistics excel files, hence, if the name is too long, the file won’t be generated.
These are followed by settings controlling the demographic sampling used to generate the virtual population. Additional PEAR sampling options are available for VBE runs based on PBPK simulations.
Parameter selection behaves similarly to the Population Simulation run mode. Lastly, there are options for creating, saving and reusing virtual populations
Runs view, Simulation Settings panel (Virtual Bioequivalence run mode: General Settings)
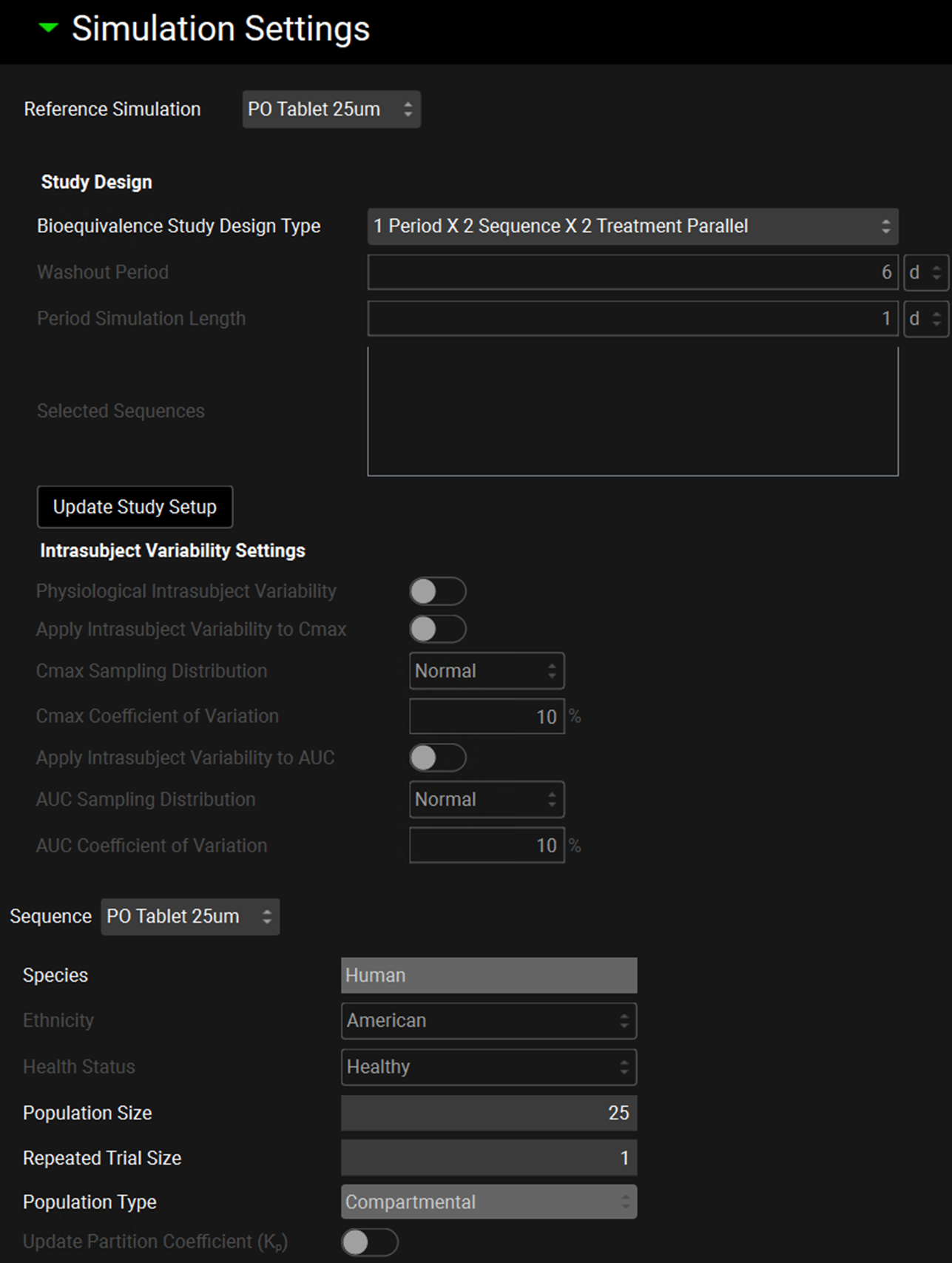
Input/Option | Description |
Reference Simulation | A drop-down. Selects which simulation (of those selected for inclusion in the current run) will be used as the reference. |
Study Design | Section where VBE study design is set up |
Bioequivalence Study Design Type | A drop-down. Selects which study design will be used in the VBE run. Bioequivalence study type may be: 1 Period x 2 Sequence x 2 Treatment Parallel* 2 Period x 1 Sequence x 2 Treatment Crossover 2 Period x 1 Sequence x 1 Treatment Fed Fasted Crossover‡ 2 Period x 2 Sequence x 2 Treatment Crossover 2 Period x 2 Sequence x 1 Treatment Fed Fasted Crossover 4 Period x 2 Sequence x 2 Treatment Crossover 3 Period x 3 Sequence x 2 Treatment Crossover *Parallel study means different virtual populations will be used in reference and test simulations. ‡Both fasted and fed simulations need to be created and selected for the VBE run before selecting this option. The bioequivalence study designs are represented in Diagram 1. |
Washout Period | Activated when replicate design is selected (2 Period x 2 Sequence or more). Determines the length of the washout period between the VBE periods. The value and the units (d or h) can be edited. |
Period Simulation Length | Activated when replicate design is selected (2 Period x 2 Sequence or more). Determines the length of each period in the selected BE study design. The value and the units (d or h) can be edited. |
Selected Sequences | Activated when replicate design is selected (2 Period x 2 Sequence or more). Determines the sequences to be used in the VBE run. The sequences displayed depend on the BE study design type selected. If the 4 Period x 2 Sequence x 2 Treatment Crossover study design is selected, the user can choose which two sequences to be used in the VBE run. For all other study designs all sequences displayed must be selected. |
Update Study Setup | A button that creates the assets needed to run the selected BE study design type. This button must be clicked in order to run replicate design VBEs (2 Period x 2 Sequence or more). Clicking this button will reset the selections, values, and CVs in the Parameter Selection section (see below). |
Intrasubject Variability Settings | Section where intrasubject variability is set up. For Parallel study design, the intrasubject variability options below will be disabled. |
Physiological Intrasubject Variability | A toggle. Controls whether additional variability will be applied to gut physiological parameters (transit times, intestinal fluid volume, pH values, and bile salt concentrations for each gut section) in the same subject between simulations. |
Apply Intrasubject Variability to Cmax | A toggle. Controls whether post-hoc variability will be applied to Cmax after the simulations have run. This will add variability to simulated Cmax for BE calculations using a CV% value. |
Cmax Sampling Distribution | A drop-down. Selects the distribution type (Normal or LogNormal) that will be used in sampling intrasubject variability for Cmax. This is enabled only if Apply Intrasubject Variability to Cmax is toggled on. |
Cmax Coefficient of Variation | The coefficient of variation, as a percentage, that will be used in applying intrasubject variability for Cmax. This is enabled only if Apply Intrasubject Variability to Cmax is toggled on. |
Apply Intrasubject Variability to AUC | A toggle. Controls whether post-hoc variability will be applied to AUC after the simulations have run. This will add variability to simulated AUC for BE calculations using a CV% value. |
AUC Sampling Distribution | A drop-down. Selects the distribution type (Normal or LogNormal) that will be used in sampling intrasubject variability for AUC. This is enabled only if Apply Intrasubject Variability to AUC is toggled on. |
AUC Coefficient of Variation | The coefficient of variation, as a percentage, that will be used in applying intrasubject variability for AUC. This is enabled only if Apply Intrasubject Variability to AUC is toggled on. |
Sequence | A Drop-down. Selects the simulation to which the below settings will be applied. The options are populated automatically from the baseline simulations selected for VBE Run/ selected BE study design. Includes all sequences in the VBE run. Parameter sampling settings need to be setup separately for each simulation, as dictated in the parameter selection section below. Users should ensure that the parameters selected for sampling and the respective ranges align between the test and reference simulations. |
Species | The species of which the virtual population will be comprised. This is based on the simulation selected for the run and cannot be edited here. |
Ethnicity | A drop-down. Selects the ethnicity to be sampled in the population: American, Chinese, Japanese. Affects sampling of demographic information, as well as additional PEAR parameters for PBPK simulations. Initial value is based on the physiology schedule in the baseline simulation. The selection can be edited. |
Health Status | A drop-down. Selects the health status of the sampled population, Cirrhosis A, B, C, Healthy, Morbidly Obese, NonAlcoholic Fatty Liver Disease, NonAlcoholic Steato Hepatitis, Obese, Pregnant, Renal Impair End Stage, Renal Impair Mild, Renal Impair Moderate, Renal Impair Severe. Affects PEAR parameters for PBPK simulations. Initial value is based on the physiology schedule in the baseline simulation. The selection can be edited. |
Population Size | The number of subjects to include in the virtual population. The value can be edited. Note: If a population is selected in the Generate Populations Settings table below, this selection will be disabled. |
Repeated Trial Size | The number of repeat trials to be conducted. Each additional trial will resample a new population within the same parameter ranges. The value can be edited. Note: If a population is selected in the Generate Populations Settings table below, this selection will be disabled. |
Population Type | A drop-down. Selects which population sampling method to use. If the baseline simulation uses a compartmental PK model, only compartmental populations will be available. If the baseline simulation is a PBPK simulation, the options are PEAR or Monte Carlo, with PEAR being the default. |
Update Partition Coefficient | A toggle. Controls whether tissue:plasma partition coefficients (Kps) will be updated during the VBE run. This is relevant if parameters used in the calculation of Kps (e.g. pKa, LogD, Fup, Rbp) are varied. This option is enabled only for simulations using a PBPK model. Note: If this toggle is ON and the Kps are also selected for sampling, the sampled values will get overwritten by the calculated Kp. This value can be seen by outputting the Inputs. |
Diagram 1: Schematic diagram of Bioequivalence Study Designs
1 Period x 2 Sequence x 2 Treatment Parallel
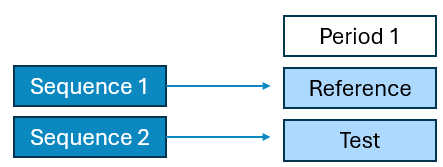
2 Period x 1 Sequence x 2 Treatment Crossover
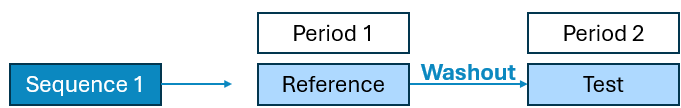
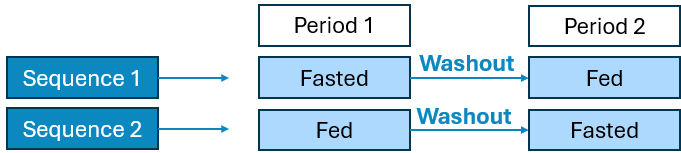
2 Period x 1 Sequence x 1 Treatment Fed Fasted Crossover

2 Period x 2 Sequence x 2 Treatment Crossover
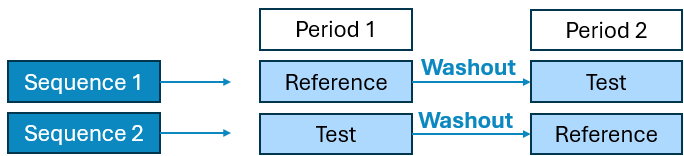
2 Period x 2 Sequence x 1 Treatment Fed Fasted Crossover
4 Period x 2 Sequence x 2 Treatment Crossover
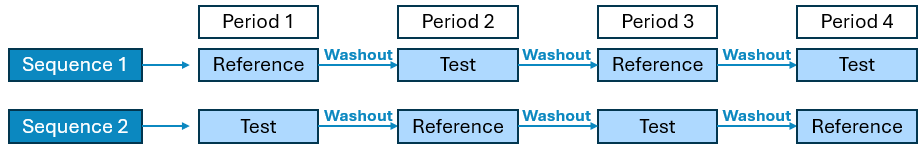
Designs available:
R-T-R-T / T-R-T-R
R-R-T-T / T-T-R-R
R-T-T-R / T-R-R-T
3 Period x 3 Sequence x 2 Treatment Crossover
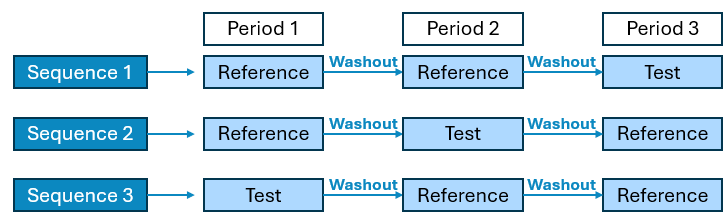
Human PEAR Settings Sub-panel
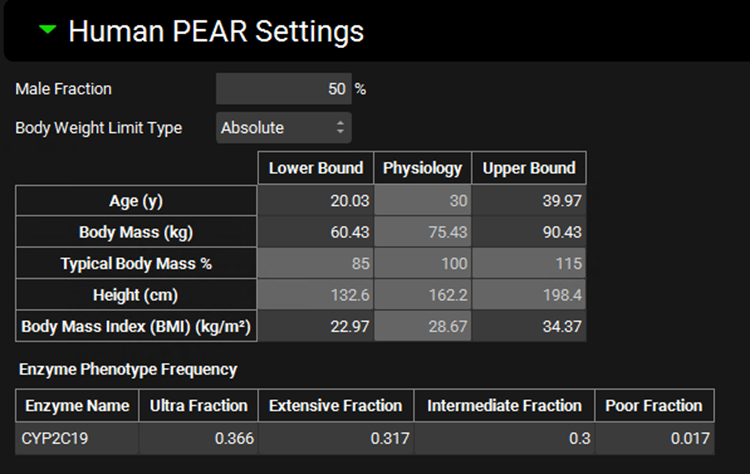
Once the initial settings have been set, additional options are available in the Human PEAR Settings sub-panel, where users can specify the percentage of the population that is male and the baseline PEAR physiology sampling ranges for Age, Weight, Height, Body Mass Index (BMI), and Enzyme Phenotype Frequency. These settings are only available for human PBPK simulations.
Runs view, Simulation Settings panel (Virtual Bioequivalence run mode), Human PEAR Settings sub-panel
Input/Option | Description |
Male Fraction | The proportion of subjects, as a percentage, of the virtual population that will be male. The remainder will be female. The value can be edited. |
Body Weight Limit Type | Selects whether sampling of body mass will be bounded by absolute numbers or by percentage of baseline body mass. This will control which of those range options is enabled below. |
Age | The range for sampling the age, in years, of subjects in the virtual population. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is provided for informational purposes only. |
Body Mass | The range for sampling body mass, in kg, of subjects in the virtual population. This option is enabled only if Body Weight Limit Type is set to Absolute. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is provided for informational purposes only. |
Typical Body Mass % | The range for sampling body mass, as a percentage of baseline, of subjects in the virtual population. This option is enabled only if Body Weight Limit Type is set to Percentage. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is used to determine the numerical bounds based on the percentages entered. |
Height | The range for sampling the height, in cm, of subjects in the virtual population. This value is calculated from body mass and BMI and changes in either body mass or BMI will cause Height to be automatically recalculated. It cannot be edited and is provided for information only. |
Body Mass Index (BMI) | The range for sampling the BMI, in kg/m2, of subjects in the virtual population. The lower and upper bounds can be edited. The baseline value (shown in the physiology column) is derived from the physiology in the baseline simulation and is provided for informational purposes only. |
Enzyme Phenotype Frequency | This section controls the relative frequency of phenotype occurrence for relevant enzymes in the virtual population. The actual expression of each enzyme will still be sampled (if selected as a parameter for sampling below), but the baseline value from which the sampling is done will be based on the phenotype. The chance of a given subject having a given phenotype is set here. This section will only be active for enzymes active in the baseline simulation which have kinetics defined for two or more phenotypes in the enzyme kinetics table. |
Enzyme Name | The name of the enzyme for which the phenotype frequencies will apply. |
Ultra, Extensive, Intermediate, and Poor Fractions | The relative frequencies, as fractions, of the respective enzyme phenotypes. The default values are based on available literature information for enzyme phenotype distributions in the given ethnicity. These values can be edited but must add up to one. |
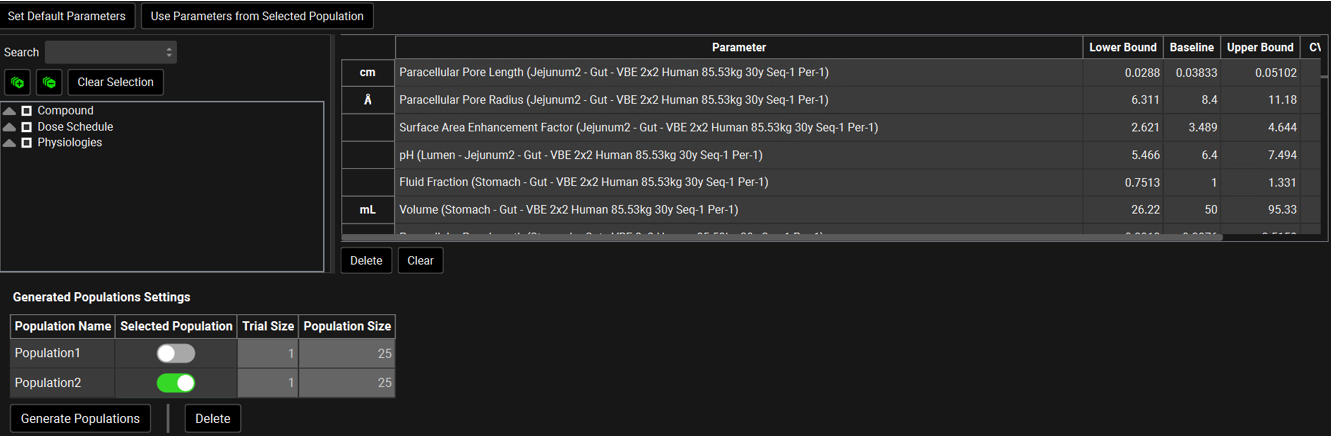
Virtual Bioequivalence Parameter Selection
The parameter selection tree and table behave similarly to the PSA, Population Simulation, and Optimization run modes. As with Population Simulation Runs, a list of parameters is selected by default for VBE, based on the parameters expected to vary in a population. The baseline values for these parameters are derived from the selected simulation. The coefficient of variation (CV) values are based on literature data where available, with the default being 10% for the remainder. This CV is applied to the baseline value from the baseline simulation using the indicated distribution type to sample the parameter value that will be applied. A new sampling is done for each subject to achieve the desired distribution.
Of note, for PEAR populations, physiological parameters calculated based on the PEAR physiology will have the “PEAR” distribution type available and selected by default. This method first recalculates the baseline value based on the sampled subject’s PEAR physiology (see Human PEAR Sampling above). Sampling is then conducted for each subject using their unique baseline value and a log-normal distribution type, utilizing the indicated CV. This ensures that sampling of physiological parameters aligns with the subject’s demographic information.
Runs view, Simulation Settings panel (Virtual Bioequivalence: Parameter Selection)
Input/Option | Description |
Set Default Parameters | Resets the parameter selection and values, CV %, Bounds Factor and Distribution to the GastroPlus® defaults. |
Use Parameters from Selected Population | Resets the parameters table to reflect the population selected in the Generated Populations Settings table below. |
Parameter Selection Tree | A hierarchy tree for selecting parameters for sampling. Individual parameters can be identified by expanding and browsing the tree, or by using the search bar at the top. Parameters are selected/deselected using the check boxes. Once selected, parameters will appear in the table on the right. The available parameters are determined based on the inputs to the simulation(s) selected for the run. A variety of relevant parameters are already selected by default for VBE runs. This tree selector functions the same way in PSA, Population Simulation, Optimization, and VBE Run modes. |
Search | A text entry field. Used to search the parameter selection tree for parameters by name. To reset the parameter selection tree, clear the text in the search box and press Enter. |
[+] and [-] buttons | Expand or collapse the entire parameter selection tree. |
Clear Selection | Unchecks the checkboxes for all parameters and clears the table to the right. |
Parameter | The name of the parameter(s) being sampled in the virtual bioequivalence run. |
Lower Bound | The lower bound of the sampling range, as determined by the baseline value, the CV, and the distribution type. This value is provided for informational purposes only; it cannot be edited. |
Baseline | The baseline value for the parameter, derived from the baseline simulation. This value can be edited to override the default baseline. Note that for parameters using the PEAR Sampling distribution type editing this value will have no effect as this value is recalculated internally for each subject based on their sampled demographic information. |
Upper Bound | The upper bound of the sampling range, as determined by the baseline value, the CV, and the distribution type. This value is provided for informational purposes only; it cannot be edited. |
CV | The coefficient of variation, in percent, of the parameter distribution. Default values are based on literature information where available; other parameters have a default of 10%. This value can be edited. Editing this value will trigger a recalculation of the Std Dev and the Lower and Upper Bounds. This value will be automatically recalculated if Std Dev is edited. |
Std Dev | The standard deviation of the parameter distribution. The default value is calculated based on the default CV. This value can be edited. Editing this value will trigger a recalculation of the CV and the Lower and Upper Bounds. This value will be automatically recalculated if CV is edited. |
Bounds Factor | The number of standard deviations describing the sampling range. This value affects the Lower and Upper Bounds and can be edited. |
Distribution | Selects the distribution type for sampling. Options include Uniform, Normal, Log-Normal, and, if applicable, PEAR. The default selection for all non-PEAR parameters is Log-Normal. |
Crossover | A toggle that indicates whether the parameter will be resampled for the second period simulation or reused from the first period population. If the toggle is off, the value sampled in the population generated in the first period simulation will be reused. If on, the parameter will be resampled. |
Regimen Scaling | A toggle that indicates gut physiological parameters (transit times, intestinal fluid volume, pH values, and bile salt concentrations for each gut section) that will be scaled in the second period. |
Delete | Deletes the selected parameter from the table and deselects the parameter in the parameter selection tree on the left. |
Clear | Deletes all parameters from the table and deselects all parameters in the parameter selection tree on the left. |
Generated Populations Settings | Section where populations are generated |
Population Name | Name given by the user for the generated population. |
Selected Population | A toggle. Indicates which population will be used in the run. |
Trial Size | The number of repeat trials to be conducted in the run. This value is set up in the “Repeated Trial Size” field above. |
Population Size | The number of subjects included in the virtual population. This value is set up in the “Population Size” field above. |
Generate Populations | A button that creates a population based on the previous selections. |
Delete | A button. Deletes the highlighted population. |
Using “User Defined Output Points” in the simulations included in the VBE run is a best practice to avoid differences in AUC, Cmax and Tmax between runs using the same population. This setting is found under the Configuration panel in the Simulations view.
When multiple Forms are present in the selected simulations and the Forms fractions are selected to be permutated, the iteration file will list the values generated by the run mode. These fractions assigned by the run are then normalized in the engine when the simulation is executed. The normalized values can be seen by outputting the Inputs.
Output Settings Panel
The Output Settings Panel provides control over what output formats, if any, will be generated after the run completes. Both simulation inputs and outputs, including summary statistics and detailed results, are available in multiple formats.
Output files are saved to the same directory as the project files, within a folder matching the run name.
The settings here apply to all run types, although optimization runs have more limited options.
Runs view, Output Settings panel
Runs view, Output Settings panel for Optimization Runs
Input/Option | Description |
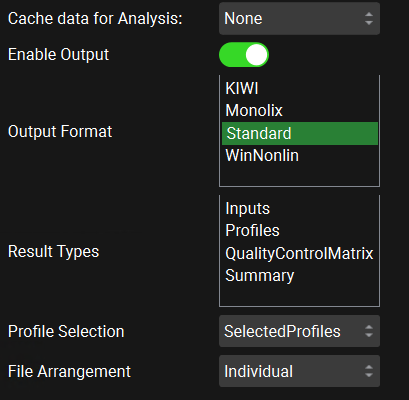
Cache data for Analysis | Selects how much data from simulation results is saved for display within the GPX™ interface. The default selection for Simulations, PSA and Optimization is All. The default selection for Population Simulations and VBE is Minimal. This option does not affect what results will be written to the output files. This option can be changed to Minimal or None to conserve memory when a run contains a large number of simulations, such as population simulations with large population sizes. The Minimal option will save the plasma concentration series along with summary absorption series and statistics. None will not save any results for view in the interface. |
Enable Output | A toggle. Controls whether one or more output files will be generated based on the settings below. |
Output Format | Selects which format(s) will be used for the output file(s). Multiple formats may be selected, each being written to a separate excel file. KIWI, Monolix, and Winnolin formats are compatible with the stated software. |
Result Types | Selects which types of results to include in the output. |
Profile Selection | Selects whether to write all profiles to the output, or only selected profiles. If Selected Profiles is selected, options for selecting those profiles will be enabled below. Selected Profiles is selected by default. |
File Arrangement | Selects whether to write out the results for each simulation in separate files, or in a composite file, with each simulation having a separate worksheet or section. Individual is selected by default. Composite file arrangement is not compatible with outputting profiles if the Output Format is Standard. If the Output Format is Winnolin, only the Cp-time profiles will be outputted, regardless of the profiles selected. Individual file arrangement is compatible with outputting either Selected or All Profiles, regardless of the Output Format. |
If Profile Selection is set to Selected Profiles, further options are enabled to select the desired series for each simulation.
Runs view, Output Settings panel (Selected Profiles option)
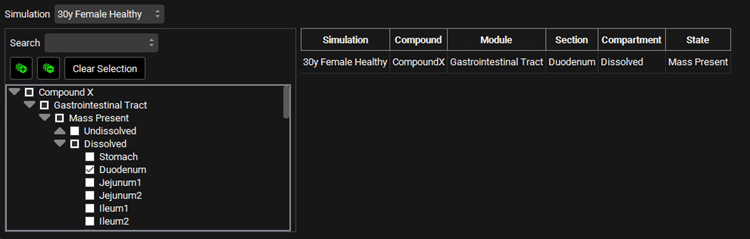
Input/Option | Description |
Simulation | Selects the simulation from which the output profile will be taken. Individual selections must be made for each simulation. The search remains active when the Simulation is switched resulting in easy selection of the same profiles for a number of simulations. |
Profile Selection Tree | A hierarchy tree for selecting profiles for output. Individual profiles can be identified by expanding and browsing the tree, or by using the search bar at the top. Profiles are selected/deselected using the check boxes. Once selected, profiles will appear in the table on the right. The available profiles are determined based on the simulation(s) selected for the run. This tree selector functions similarly to other parameter selection trees in GPX™, though the options here are specific to simulation outputs. |
Search | A text entry field. Used to search the profile selection tree for profiles by name. To reset the parameter tree, clear the text in the search box and press Enter. |
[+] and [-] buttons | Expand or collapse the entire profile selection tree. |
Clear Selection | Unchecks the checkboxes for all profiles for the selected simulation. Will also clear the table to the right. |
Simulation, Compound, Module, Section, Compartment, and State | Provides a summary of the series selected, as identified by location, tissue, and series type information, for each compound in each simulation. |
Run Controls Panel
The Run Controls Panel provides options for starting and stopping runs, as well as providing feedback to the user about the status of the run.
Runs view, Run Controls Panel

Input/Option | Description |
Start | Starts the run. |
Stop | Stops the run. Only applicable after a run has been started. |
Most Recent Run Output | Provides feedback to the user about the status/progress of a run. For most run types, this will provide information about the progress of the run in terms of starting and completion. For optimization runs, this will provide information about the status of the optimization (current parameter values, step size, objective function value, etc.). |
