ADMET Predictor® Module
The ADMET Predictor® Module is used to import compounds into a GPX™ project, with options for populating the Compound, Dosing, and Pharmacokinetic asset properties. Physicochemical and pharmacokinetic properties of the compound(s) may be predicted using the ADMET Predictor® models, or, along with dosing information, filled with user-provided values. The ADMET Predictor® Module will also automatically create a simulation in the GPX™ project for each compound imported, allowing for quick running and comparison of simulations based on ADMET Predictor® imports.
The ADMET Predictor® Module takes a molecular structure file (.smi, .mol, .mol2, .sdf, etc.) as input, which contains structural data for one or more discrete molecules. If additional properties have been measured for these compounds, and the chosen file type allows it, those may also be included for each compound in the file. When importing the structure, users will have the option to let ADMET Predictor® predict physicochemical and pharmacokinetic properties of the compound(s), in some cases selecting additional options regarding which predictive models to use, or to fill in specific properties with user-provided data in the structure file.
To import a compound into a GPX™ project via ADMET Predictor®, the project must first have at least one Dosing Schedule and one Physiology Schedule. The reasons for this are twofold. First, several pharmacokinetic predictions (clearance, fraction unbound in plasma, blood to plasma concentration ratio, etc.) are species specific, and thus the species of the physiology in the physiology schedule is used to determine which ADMET models to use (predictions for other species are still done and are saved in the Lab Book). Second, the ADMET Predictor® import will automatically create a simulation in the GPX™ project for each compound, allowing for quick running and comparisons of simulations based on ADMET Predictor®. As such, ADMET Predictor® imports require a Dosing Schedule asset and a Physiology Schedule asset with which to build the simulation.
The ADMET Predictor® Module consists of three panels. The General Settings panel, where the structure file, Dosing and Physiology Schedules, and clearance models can be selected. The Advanced Settings panel enables more detailed control over the remaining properties. Lastly, the Import panel enables users to complete the ADMET Predictor® Import and provides information on the imported compound(s) once the import has completed.
General Settings
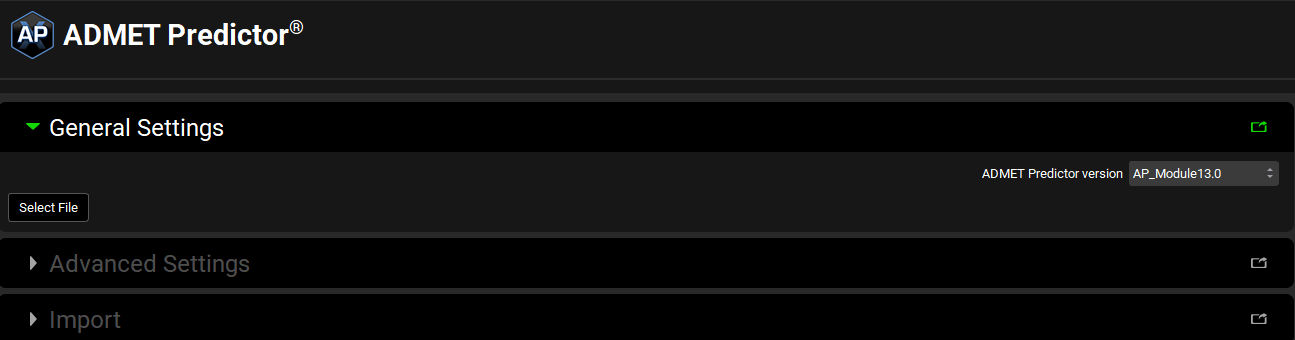
The General Settings panel first enables selection of a structure file for import. Until a structure file is selected, no other options will be available. Once a file has been selected, additional options will become available.
ADMET Predictor® Module, General Settings panel (no structure selected)
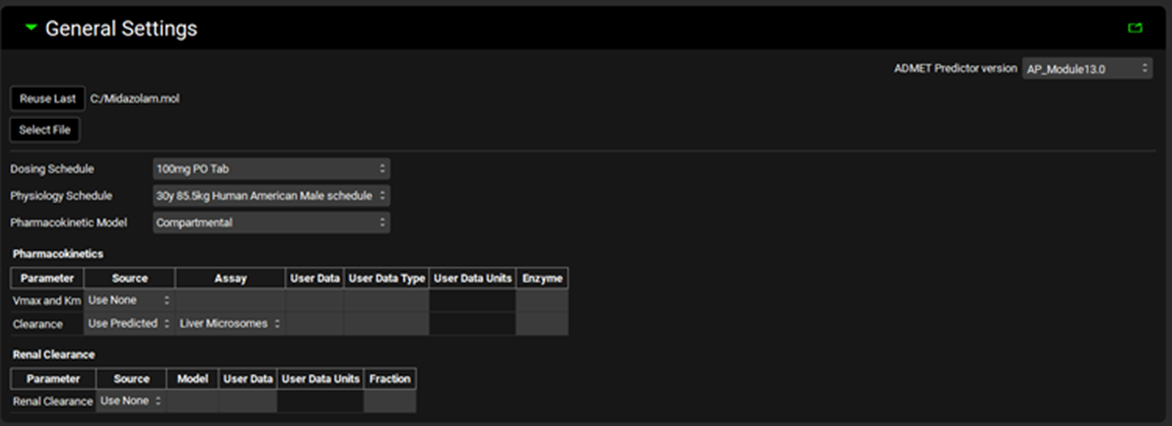
ADMET Predictor® Module, General Settings panel (structure selected)
Input/Option | Description |
Select File | Opens the windows explorer dialogue, enabling selection of the structure file for import. |
Reuse Last | Reuses the last imported file (the filename and path are displayed next to the button). |
ADMET Predictor® version | A drop-down. Selects the version of ADMET Predictor® to use for the import which will affect which models are used in the predictions. Options are populated based on the versions installed. The updated version for GPX™ 10.2 is AP_Module 13.0. |
Dosing Schedule | A drop-down. Selects the Dosing Schedule to use for the import. Options are populated from Dosing Schedules already added to the projects. A Dosing Schedule must be selected to enable below options and to proceed with the import. |
Physiology Schedule | A drop-down. Selects the Physiology Schedule to use for the import. Options are populated from Physiology Schedules already added to the projects. A Physiology Schedule must be selected to enable below options and to proceed with the import. |
Pharmacokinetic Model | A drop-down. Selects the type of pharmacokinetic model (Compartmental or PhysiologicallyBased) to use for the import. The option selected here will determine the type (compartmental or PBPK) of the simulations created by ADMET Predictor®, and will also affect the in vivo clearance predictions. This selection here will not affect the results of physicochemical property prediction. If the Pharmacokinetic Model is set to PhysiologicallyBased, additional options will be enabled for Renal Clearance and for Kp calculations. Additionally, if a PEAR physiology has not yet been setup, one will be created upon import based on the demographics of the physiology schedule selected above. |
Pharmacokinetics Table | This section controls settings for metabolic clearance predictions. |
Parameter | The parameter to which the remaining information in the table applies. In this case, there are only two mutually exclusive entries:
Only one of these options may be selected, as indicated in the Source column to the right (see below). Note: Some of these predictions may not be available dependent on the licenses purchased. |
Source | The source of the parameter value:
Only Vmax and Km OR Clearance may have a source selected, the other must be Use None. GPX™ will automatically set one to Use None when Use Predicted is set for the other. |
Assay | Which predictive models, based on assay type, will be used to predict the clearance.
This column is enabled only if the Source is set to Use Predicted. |
User Data | Selects the field in the structure file that corresponds to the indicated parameter. Options are populated based on columns/entries in the structure file. This column is only available for Clearance, not Vmax and Km, and is enabled only if the Source is set to User Data. |
User Data Type | Selects the clearance measurement type, InVitro or PerGramTissue, which is used in the in vitro to in vivo conversion. This column is only available for Clearance, not Vmax and Km, and is enabled only if the Source is set to User Data. |
User Data Units | Selects the units for the user-provided data value ((mL/min)/g, (µL/min)/g, (µL/s)/g, (mL/s)/g. This column is only available for Clearance, not Vmax and Km, and is enabled only if the Source is set to User Data. |
Enzyme | Not currently enabled |
Renal Clearance Table | This section controls settings for renal clearance predictions. |
Parameter | The parameter to which the remaining information in the table applies. In this case, there is only one entry: Renal Clearance. |
Source | The source of the parameter value:
If the Pharmacokinetic Model is set to compartmental, only User Data and Use None will be available. If the Pharmacokinetic Model is set to PhysiologicallyBased, Use Predicted will also be available as a source. The method of prediction is then controlled in the Model column. |
Model | Selects the renal clearance model to use. Available only for Physiologically Based imports when the Source is set to Use Predicted. Options are:
|
User Data | Selects the field in the structure file that corresponds to the indicated parameter. Options are populated based on columns/entries in the structure file. This column is enabled only if Source is set to User Data. |
User Data Units | Selects the units for the user-provided data value. This column is enabled only if Source is set to User Data. |
Fraction | The fraction of kidney blood flow that represents renal clearance. For use when Model is set to Fraction Blood Flow. |
Compartmental and PBPK models can exist side-by-side in GPX™ for any given compound-physiology pair. As such, physiologically based ADMET Predictor® imports will create both a PBPK and a compartmental pharmacokinetic model.
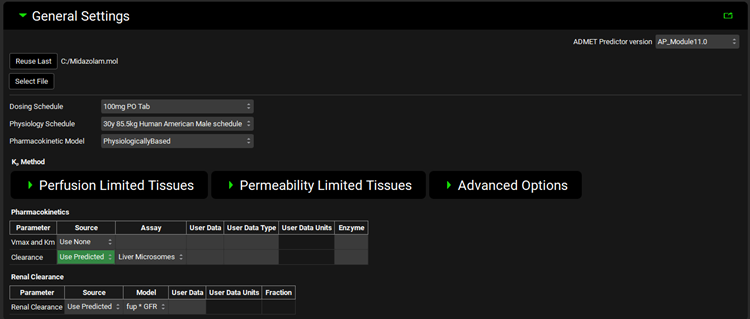
When the Pharmacokinetic Model is set to PhysiologicallyBased, additional options become available for Kp calculation settings. These settings are identical to those on the PBPK panel (See PBPKPlus™ panel).
ADMET Predictor® Module, General Settings panel (structure selected, Physiologically Based Pharmacokinetic Model selected)
Advanced Settings
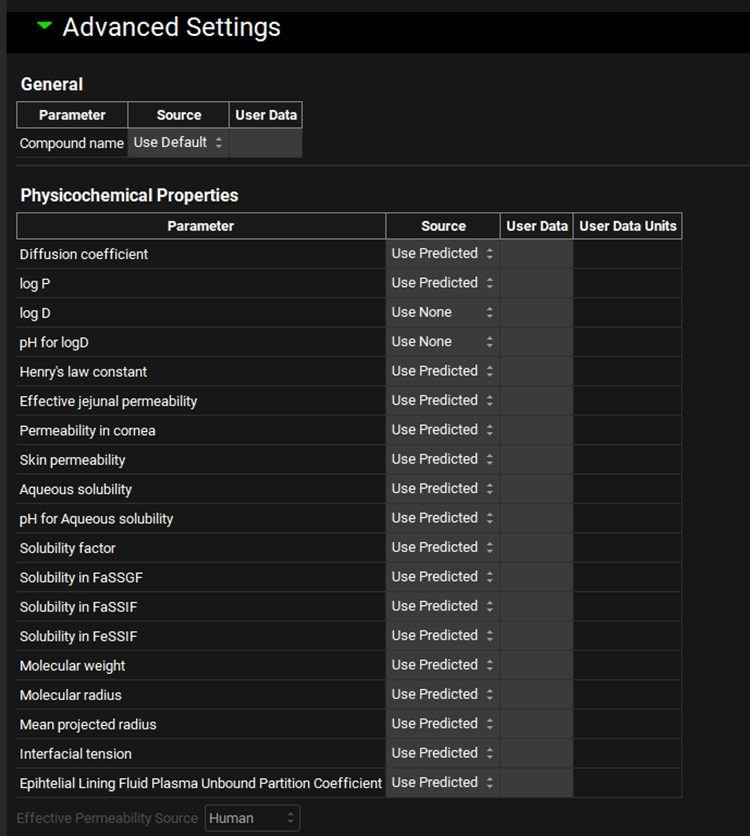
The Advanced Settings panel enables further mapping of User Data for additional compound, dosing, and pharmacokinetic properties. By default, all compound parameters are set to Use Predicted, meaning ADMET Predictor® will predict the values based on the structure and assign them to the compound asset upon import. Any of these can optionally use User Data instead.
ADMET Predictor® Module, Advanced Settings panel (General and Physiochemical Properties)
Input/Option | Description |
General | Covers general information about the compound, such as the Compound Name. |
Parameter | The parameter to which the remaining information in the table applies. In this case, there is only one entry: Compound Name. |
Source | The source of the parameter value:
|
User Data | Selects the field in the structure file that corresponds to the compound name. Enabled only if Source is set to User Data. |
Physicochemical Properties | Covers import settings for physiochemical properties of the compound. |
Parameter | The parameter to which the remaining information in the table applies. |
Source | The source of the parameter value:
|
User Data | Selects the field in the structure file that corresponds to the indicated parameter. Enabled only if Source is set to User Data. |
User Data Units | Selects the units for the user-supplied value, where applicable. Enabled only if Source is set to User Data. Note: Please check that the units in the structure file are accepted by GPX™. |
Effective Permeability Source | A drop-down. Selects the source for the Effective Jejunal Permeability measurement. Applies only if the Source for Effective Jejunal Permeability is set to User Data. |
Formulation and dosing parameters will, by default, be based on the Dosing Schedule selected above (“Use Default” option). However, any of these can be substituted with User Data from the structure file if desired. These dosing parameters are used to create additional Dose Schedules that will be used in the simulations automatically created by ADMET Predictor® for each compound upon import. They will then be available for use within the current project in any user-defined Simulation.
Similarly, the Observed Values will be set to Use None by default, but User Data can be supplied as needed. The imported values will be available on the Observed Data view on the Parameters Panel. Physiological and pharmacokinetic parameters, as with compound properties, will be predicted by default, but may be substituted with User Data.
ADMET Predictor® Module, Advanced Settings panel (Formulation Parameters, Observed Values, Physiology and Pharmacokinetics, and Clearance)
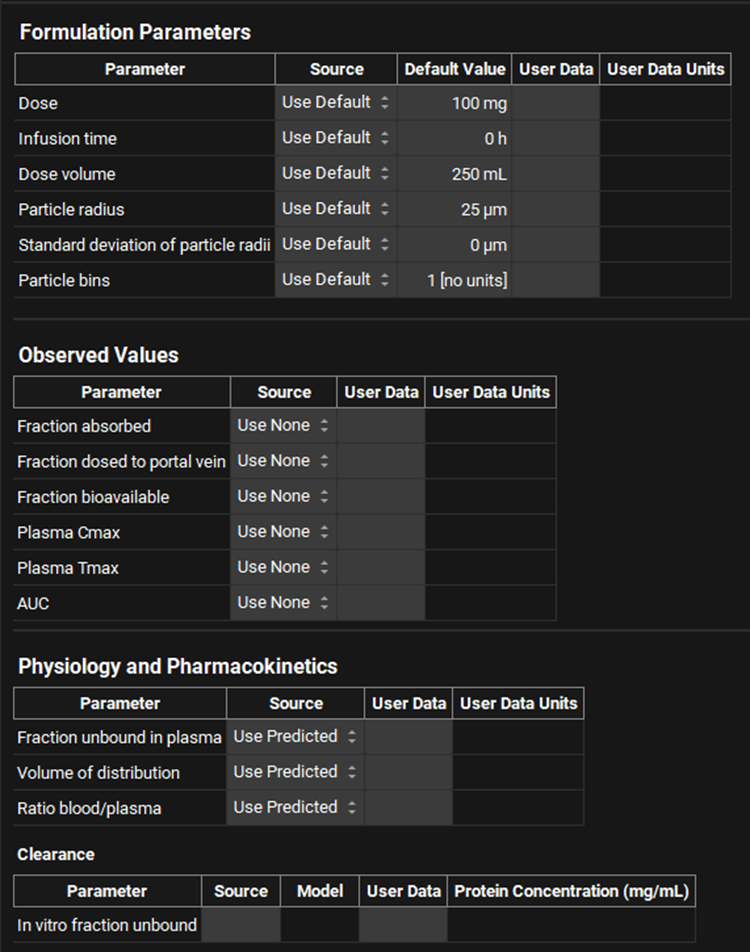
Input/Option | Description |
Formulation Parameters | This table controls updates to the default Dosing Schedule (selected under General Settings). |
Parameter | The parameter to which the remaining information in the table applies. |
Source | The source of the parameter value:
|
Default Value | Previews the values from the default Dosing Schedule (selected under General Settings). This is shown only if Source is set to Use Default and is provided for information only. |
User Data | Selects the field in the structure file that corresponds to the indicated parameter. Enabled only if Source is set to User Data. |
User Data Units | Selects the units for the user-supplied value. Enabled only if Source is set to User Data. |
Observed Values | Covers import settings for observed pharmacokinetic parameters for the compound. The values will be imported to the Parameters panel in the Observed Data view. |
Parameter | The parameter to which the remaining information in the table applies. |
Source | The source of the parameter value:
|
User Data | Selects the field in the structure file that corresponds to the indicated parameter. Enabled only if Source is set to User Data. |
User Data Units | Selects the units for the user-supplied value where applicable. Enabled only if Source is set to User Data. |
Physiology and Pharmacokinetics | Covers import settings for pharmacokinetic properties of the compound. |
Parameter | The parameter to which the remaining information in the table applies. |
Source | The source of the parameter value:
|
User Data | Selects the field in the structure file that corresponds to the indicated parameter. Enabled only if Source is set to User Data. |
User Data Units | Selects the units for the user-supplied value where applicable. Enabled only if Source is set to User Data. |
Clearance | Controls settings for predicting or importing the in vitro fraction unbound, which is used for in vitro to in vivo conversion of imported linear clearance values. This section is relevant only if the Source for Clearance (under General Settings) is User Data. |
Parameter | The parameter to which the remaining information in the table applies. In this case, in vitro fraction unbound is the only parameter. |
Source | The source of the parameter value:
|
Model | Selects the model for predicting in vitro fraction unbound from the following options: Calculated Austin HLM, Calculated Austin Hep, Calculated Hallifax HLM, Use Plasma Fu, Is Unbound. Enabled only if Source is set to Use Predicted. |
User Data | Selects the field in the structure file that corresponds to the indicated parameter. Enabled only if Source is set to User Data. |
Protein Concentration | The in vitro microsomal protein concentration, in mg/mL. This value is used in the Austin and Hallifax models. This field is enabled only if the Source is set to Use Predicted, and the Model is set to Calculated Austin HLM or Calculated Hallifax HLM. The value can be edited. |
IVIVE sub-panel
The IVIVE (in vitro in vivo extrapolation) sub-panel contains advanced settings for controlling the scaling factors and coefficients used the in vitro to in vivo clearance conversions.
ADMET Predictor® Module, Advanced Settings panel, IVIVE sub-panel
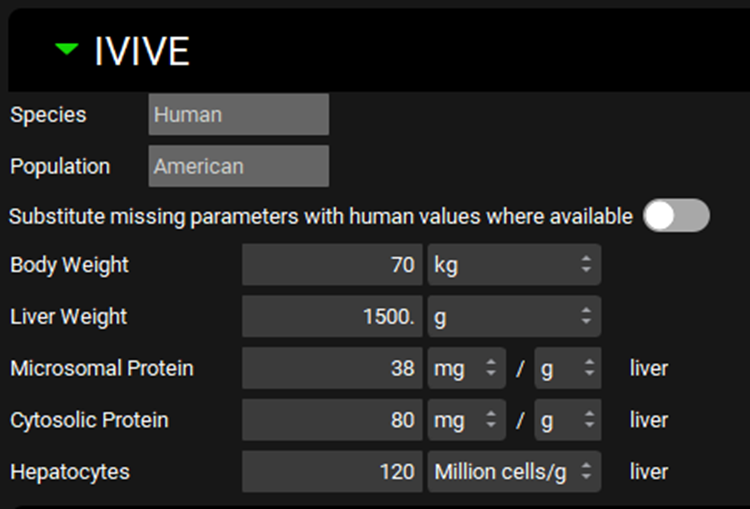
Input/Option | Description |
Species | The species for the current import. This is derived from the Physiology Schedule selected under General Settings. |
Population | The ethnicity (American, Chinese, or Japanese) for the current import. This is derived from the physiology schedule selected under General Settings. Applies only if the Species is Human. |
Substitute missing parameters with human values where applicable | A toggle. Controls whether missing parameters will use human values, if available. This is applicable when ADMET Predictor® does not have a model for a relevant parameter in a given species. For example, there is no Swine model for fraction unbound in plasma, which is a parameter used in systemic clearance calculation. If the import were to be done against a Swine physiology, this option (if toggled On) would use the predicted human fraction unbound in plasma for the clearance calculations. If toggled off, and no species-specific value exists, the default value will be used. |
Body Weight | The total body mass of the subject for which the IVIVE conversion will be done. The default value is 70 kg. The value and the units (kg, g) can be edited. If the import is Physiologically Based, this value will be ignored, and the body mass corresponding to the first physiology in the Physiology Schedule selected under General Settings will be used. |
Liver Weight | The liver mass of the subject for which the IVIVE conversion will be done. The default value is 1500 g. The value and the units (kg, g) can be edited. If the import is Physiologically Based, this value will be ignored, and the liver mass corresponding to the first physiology in the Physiology Schedule selected under General Settings will be used. |
Microsomal Protein | The mass of microsomal protein per mass liver tissue. The value and the units (mg/g, g/g, mg/kg, g/kg) can be edited. |
Cytosolic Protein | The mass of cytosolic protein per mass liver tissue. The value and the units (mg/g, g/g, mg/kg, g/kg) can be edited. |
Hepatocytes | The number of cells, in millions, per gram liver tissue. The value can be edited. |
Enzyme Expression sub-panel
The enzyme expression sub-panel, within the IVIVE sub-panel, provides additional control over enzyme expression levels, which are used in the in vitro to in vivo conversion of Vmax and linear clearance when this is predicted using the rCYP or 3A4-HLM, others-rCYP models.
ADMET Predictor® Module, Advanced Settings panel, IVIVE sub-panel, Enzyme Expression sub-panel
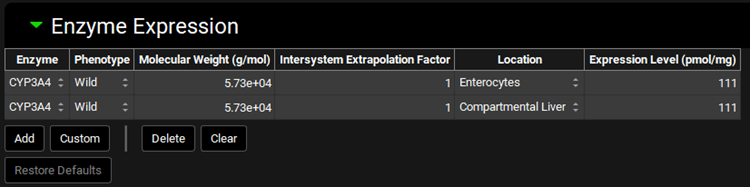
Input/Option | Description |
Enzyme | Selects the enzyme for which the remaining settings will apply. |
Phenotype | Selects the phenotype of the enzyme. |
Molecular Weight | The molecular weight, in g/mol, of the enzyme. Built-in enzymes will have this filled out by default. The value can be edited. |
Intersystem Extrapolation Factor | The intersystem extrapolation factor (ISEF) for the expression of the enzyme. The value can be edited. |
Location | The model-level location for the enzyme entry. |
Expression Level | The expression, in pmol enzyme / mg microsomal protein, of the indicated enzyme. |
Add | Adds a new set of rows to the table. Built-in enzymes expressed in multiple locations (e.g. liver and gut) will have multiple rows added, one for each location. |
Custom | Add a custom enzyme for an indicated species. Once the custom enzyme is created, it will be added automatically as a row to the table. It will also be available thereafter in other enzyme drop-downs for selection. |
Delete | Delete the selected row from the table. Hold Ctrl-key and click to highlight a row for deletion. |
Clear | Clear the entire table. |
Restore Defaults | Resets the values for built-in enzymes to the default values. |
Import panel
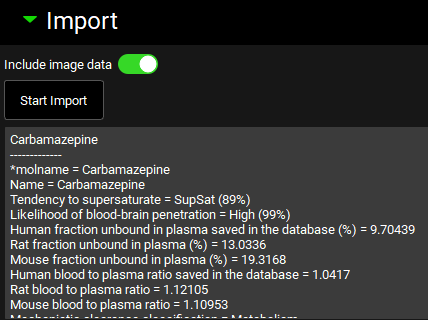
The import panel contains the start import button, which is used to conduct the ADMET Predictor® import once the above settings have been finalized. The Include image data toggle selects whether image data will be added to the project. It is on by default. If turned off, no structure image will be associated with the project. Once the import is finished, notes are visible below the Import button. These notes can also be viewed in the Lab Book View.
ADMET Predictor® Module, Import panel
ADMET Predictor® Module, Import panel, following successful import
Input/Option | Description |
Start Import | Conducts the import of the compound(s) using the ADMET Predictor® Module. |
Include image data | A toggle. On by default. Selects whether structure image data will be added to the project. If turned off, no structure image will be included in the project files. |
