DDI Module
The DDI Module is an optionally licensed module that allows predictions of competitive and/or time-dependent metabolic and transporter-based drug-drug interactions. The predictions account for the effects of any number of administered parent drugs and their metabolites formed in vivo. Both steady-state calculations (metabolic interactions only) and full dynamic simulations (metabolic and transporter-based interactions) are available.
Another optionally licensed module, the Metabolism and Transporter Module, is required to use the DDI Module.
The DDI view has seven panels which get enabled as needed depending on the selections in the first “Configuration” panel.
See:
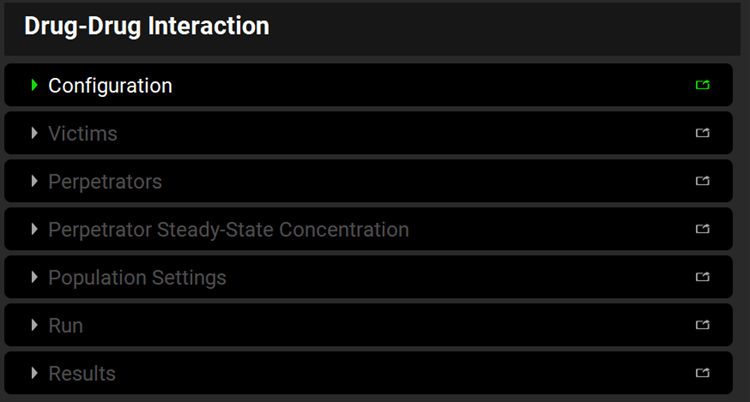
Input/Option | Description |
Configuration | Select the general settings of Physiology Schedule and Prediction Type, plus Simulation Mode for Dynamic simulations. |
Victims | Define compounds whose exposure may be impacted by co-administered drugs |
Perpetrators | Define compounds that may impact exposure of co-administered drugs |
Perpetrator Steady-State Concentration | Define effective perpetrator concentrations to use in Steady State predictions (enabled only when Steady State Prediction Type is selected in the Configuration panel). |
Population Settings | Define the population of virtual subjects to be used in the population simulation (enabled only when Dynamic Prediction Type and Population Simulation Mode is selected in the Configuration panel and after Victim and Perpetrator compounds were defined in their respective panels). |
Run | Execute DDI predictions. |
Results | View the predicted DDI results. |
Configuration panel
The general settings (i.e. Prediction Type and Physiology Schedule) are specified in this panel.
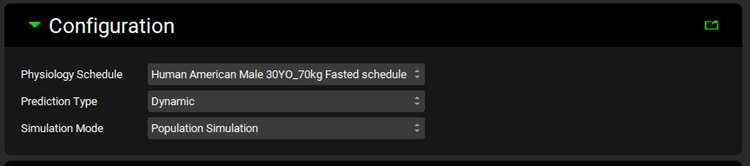
Input/Option | Description |
Physiology Schedule | Select the Physiology Schedule to use in the DDI predictions. |
Prediction Type | Select Dynamic or Steady State prediction. |
Simulation Mode | Select to use Single Simulation or Population Simulation (enabled and applicable only with Dynamic Prediction Type). |
Victims panel
Select compounds whose exposure may be impacted by co-administered compounds and the properties required for DDI predictions based on the Prediction Type selected in the Configuration panel.
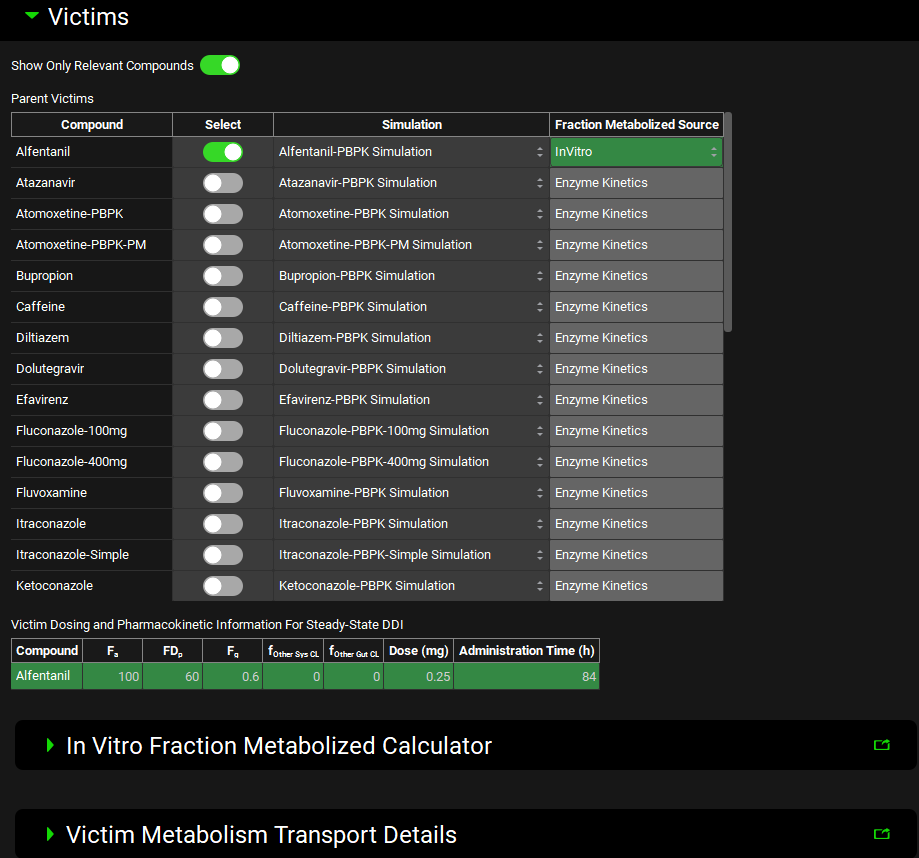
Input/Option | Description |
Show Only Relevant Compounds | Turn ON this toggle (it will turn green) to display only those compounds in the current project which have a potential for interaction with the selected Perpetrator compounds (i.e. substrates of the enzymes and transporters which are inhibited or induced by the selected Perpetrator). All compounds in the current project will be displayed if no Perpetrator compounds are selected yet. |
Parent Victims | Table of compounds in the current project that are potential victims in DDI. The list is dependent on the selections in the Configurations panel and whether the Show Only Relevant Compounds toggle is ON. When Dynamic Prediction Type is selected in the Configuration panel, the table will include only compounds that are linked to simulations with the same Pharmacokinetic Model type (Compartmental or PBPK) as defined in the Physiology Schedule selected in the Configuration Panel. The table includes up to four columns:
|
Victim Dosing and Pharmacokinetic Information for Steady-State DDI | Table summarizing the Dosing and Pharmacokinetic information of compounds selected in the Parent Victims table (applicable and enabled only with Steady State Prediction Type).
|
In Vitro Fraction Metabolized Calculator | Built-in tool for estimation of the in vivo fm values for steady-state DDI predictions from in vitro assays (in recombinant enzymes or liver microsomes). Enabled when Prediction Type in the Configuration panel is set to Steady State Prediction and the Fraction Metabolized Source for the selected Parent Victim compound is set to InVitro. |
Victim Metabolism Transport Details | Sub-panel summarizing fm values for victim compounds selected for the DDI prediction. Enabled and functional when Prediction Type in the Configuration panel is set to Steady State Prediction. To view fm for Dynamic Simulations, switch to Steady State to enable the calculations and then back to Dynamic to complete the simulation set up. |
Where units can be edited, this is done by clicking in the column header.
In Vitro Fraction Metabolized Calculator sub-panel
This sub-panel contains a tool for calculation of the in vivo fm values for steady-state DDI predictions from the in vitro clearance values measured in recombinant enzymes or liver microsomes. It is enabled only when Steady State Prediction Type is selected and at least one of the compounds selected in the Parent Victims table has Fraction Metabolized Source set to InVitro.
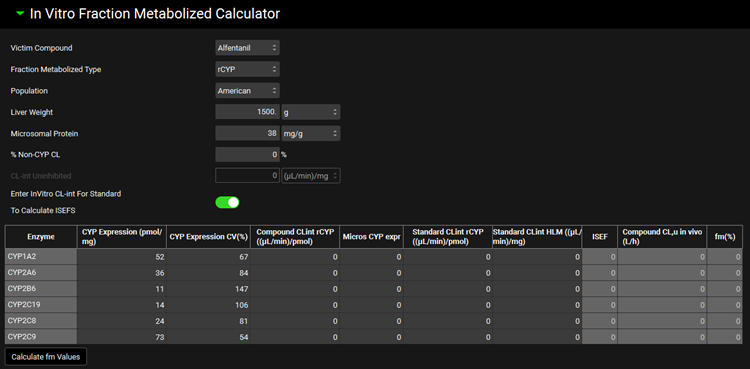
Input/Option | Description |
Victim Compound | Select the compound to calculate the fm values. The drop-down includes the list of compounds which were selected in the Parent Victims table and their Fraction Metabolized Source was set to InVitro. |
Fraction Metabolized Type | Select the type of in vitro assay (HLM for human liver microsomes or rCYP for recombinant CYP enzyme systems) that was used to determine the compound’s metabolic profile. |
Population | Select the population (American, Chinese, or Japanese) to define the average in vivo CYP expression levels in liver. |
Liver Weight | Liver weight – one of the conversion factors for extrapolation of in vitro clearance to intrinsic in vivo clearance. Default value is 1500 g. |
Microsomal Protein | Amount of microsomal protein per gram liver – a conversion factor for extrapolation of in vitro clearance to intrinsic in vivo clearance. Default value is 38 mg/g. |
% Non-CYP CL | Enter the percent of a compound’s systemic clearance (if any) due to mechanisms other than CYP metabolism. |
CL-int Uninhibited | Enter in vitro intrinsic clearance measured in HLM without the presence of any inhibitors (enabled only when HLM is selected for Fraction Metabolized Type). |
Enter InVitro CL-int For Standard To Calculate ISEFS | Turn this toggle ON to enable entries of standard compounds measured in different rCYP systems and calculate ISEFs (intersystem extrapolation factors). Turn the toggle OFF for direct entry of ISEF values. (Toggle is enabled only when rCYP is selected for Fraction Metabolized Type). |
Table for Clearance entries | In vitro measurements reflecting the compound’s metabolism by different CYP enzymes that will be used to calculate fm values:
|
Calculate fm Values | After entering all in vitro measurements in the table, click this button to calculate fm values for CYP enzymes involved in the selected compound’s elimination. The calculated fm values are automatically exported into the Victim Metabolism Transport Details panel. |
Where units can be edited, this is done by clicking in the column header.
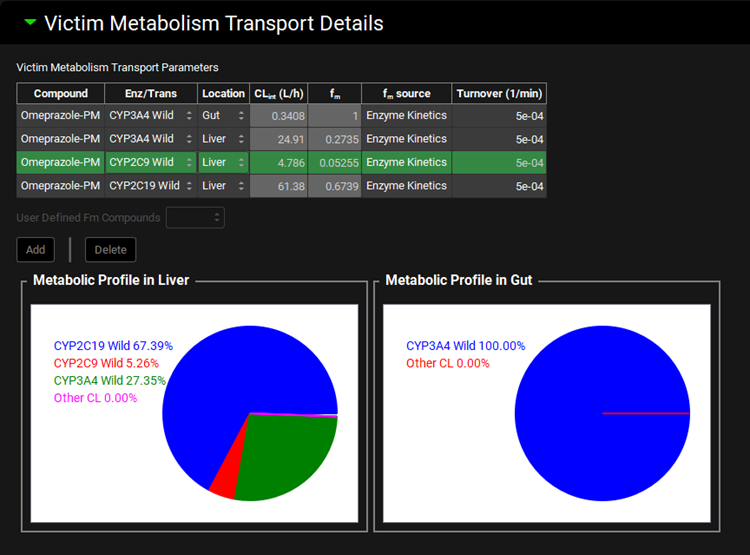
Victim Metabolism Transport Details sub-panel
This sub-panel summarizes the fm values for compounds selected in the Parent Victims table that will be used for the DDI prediction. It is active only when Steady State Prediction Type is selected. To view fm for Dynamic Simulations, switch to Steady State to enable the calculations and then back to Dynamic to complete the simulation set up.
Input/Option | Description |
Victim Metabolism Transport Parameters | Table summarizing fm values and their sources for compounds selected in the Parent Victims table:
|
User Defined Fm Compounds | A drop-down containing list of compounds selected in the Parent Victims table for which the Fraction Metabolized Source was set to User (enabled only when at least one selected Parent Victim has Fraction Metabolized Source set to User). Select compound for which a new entry needs to be added to the Victim Metabolism Transport Parameters table. |
Add | Delete | Add or Delete an entry from the table. Clicking Add button will add an entry for the compound selected in the User Defined Fm Compounds drop-down. Note: To delete an entry, select the row by holding the Ctrl key and clicking one cell in the row and then click the Delete button. These buttons are enabled only when User is selected as Fraction Metabolized Source of Parent Victims table for at least one of the selected compounds. |
Metabolic Profile in Liver/Gut | Plots of metabolic profiles in Liver and Gut for the compound selected in the Victim Metabolism Transport Parameters table (use Ctrl + click to select the row for the desired compound). |
Where units can be edited, this is done by clicking in the column header.
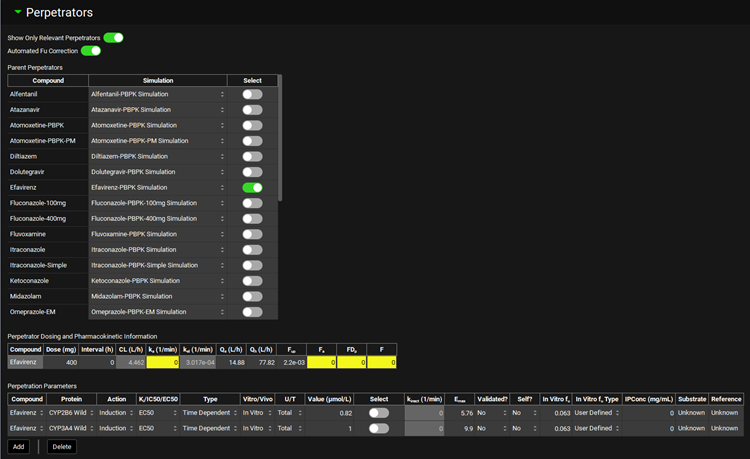
Perpetrators panel
Select compounds that may impact the exposure of co-administered compounds and the properties required for the DDI prediction based on the Prediction Type selected in the Configuration panel.
Input/Option | Description |
Show Only Relevant Perpetrators | Turn ON this toggle to display only those compounds in the current project which have a potential for interaction with the selected Victim compounds (i.e. display compounds that inhibit or induce the enzymes and transporters that impact exposure of the selected victim compounds). All compounds in the current project will be displayed if no Victim compounds are selected yet. |
Automated Fu Correction | A toggle. When turned on, it will automatically convert interaction parameter values (Ki, IC50, EC50) between the total and unbound to match the selected perpetrator concentration type. The conversion requires a defined in vitro fraction unbound in the Perpetration Parameters table. |
Parent Perpetrators | Table of compounds in the current project that are potential perpetrators in the DDI. The list is dependent on the selections in the Configurations panel and whether the Show Only Relevant Compounds toggle is ON. When Dynamic Prediction Type is selected in the Configuration Panel, the table will include only compounds that are linked to simulations with the same Pharmacokinetic Model type (Compartmental or PBPK) as defined in the Physiology Schedule selected in the Configuration Panel. The table includes three columns:
|
Perpetrator Dosing and Pharmacokinetic Information | Table summarizing the Dosing and Pharmacokinetic information of compounds selected in the Parent Perpetrators table. These values are used to provide estimates of Calculated Perpetrator concentrations in Steady State DDI predictions.
Fa, FDp and F all have to be entered in the Observed Data view, Parameters sub-panel, and the series must be linked in the Simulation view, Compound Settings panel, Observed Data sub-panel, of the relevant simulation. See Parameters Panel and Observed Data sub-panel for more details. |
Perpetration Parameters | Table summarizing interaction parameters for perpetrators used in the DDI prediction (applicable with both Steady State and Dynamic predictions).
|
Add / Delete | Add or Delete entries from the table. Note: To delete an entry, select the row by holding the Ctrl key and clicking one cell in the row and then click the Delete button. |
Where units can be edited, this is done by clicking in the column header.
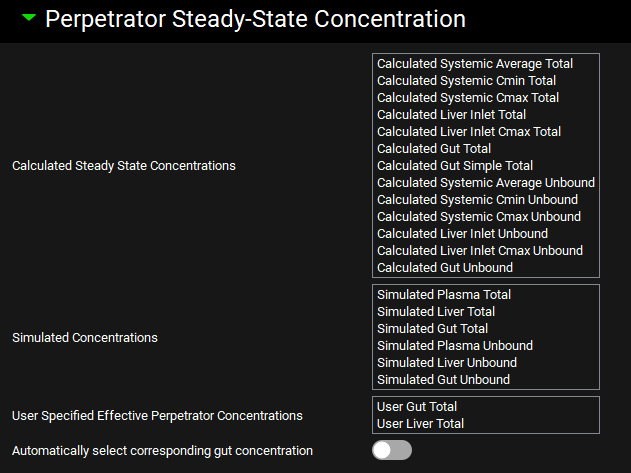
Perpetrator Steady-State Concentration sub-panel
This sub-panel provides selections of different types of perpetrator Steady-State concentrations. It is enabled only when Steady State Prediction Type is selected in the Configuration panel.
Input/Option | Description |
Calculated Steady State Concentrations | Effective perpetrator concentrations are estimated from information in the Perpetrator Dosing and Pharmacokinetic Information table using standard formulas for average, peak (Cmax) and trough (Cmin) plasma concentrations in the steady state, average and peak (Cmax) liver inlet concentrations, or drug concentration in the gut. Refer to Scientific Principles Guide or Equations Guide for the details of the equations. |
Simulated Concentrations | Selecting any of the Simulated Concentrations will cause the program to run a simulation for the perpetrator based on the information in the Perpetrator Dosing and Pharmacokinetics Information table and the settings for the Simulation selected for given perpetrator in Parent Perpetrators table. The perpetrator concentration to be used in the steady-state equation is then obtained from the simulated perpetrator concentration in plasma or liver (depending on the selection) and in the gut (if at least one of the victim’s metabolizing enzymes is located in gut). Refer to Scientific Principles Guide for details. |
User Specified Effective Perpetrator Concentrations | The User Specified Effective Perpetrator Concentrations will create an entry in the Perpetrator Steady-State Concentrations table in the Results section of the DDI window where you can manually enter the expected effective perpetrator concentration in the gut or liver. *** Keep in mind that, unlike Calculated or Simulated Perpetrator concentrations, these User Specified Effective Perpetrator Concentrations are fixed numbers. The program will not scale them for a different perpetrator dose or dosing interval. It is user’s responsibility to ensure that the value corresponds to the intended dosing of the perpetrator. |
Automatically select corresponding gut concentration | When the Automatically select corresponding gut concentration toggle is turned ON, the steady-state DDI predictions will be calculated by combining total systemic with total gut and unbound systemic with unbound gut concentrations within each section (using ‘calculated’ gut concentration with ‘calculated’ plasma or liver concentration and ‘simulated’ gut concentration with ‘simulated’ plasma or liver concentration). Otherwise, the module will produce steady-state predictions for all combinations of the selected systemic/liver and gut concentrations. |
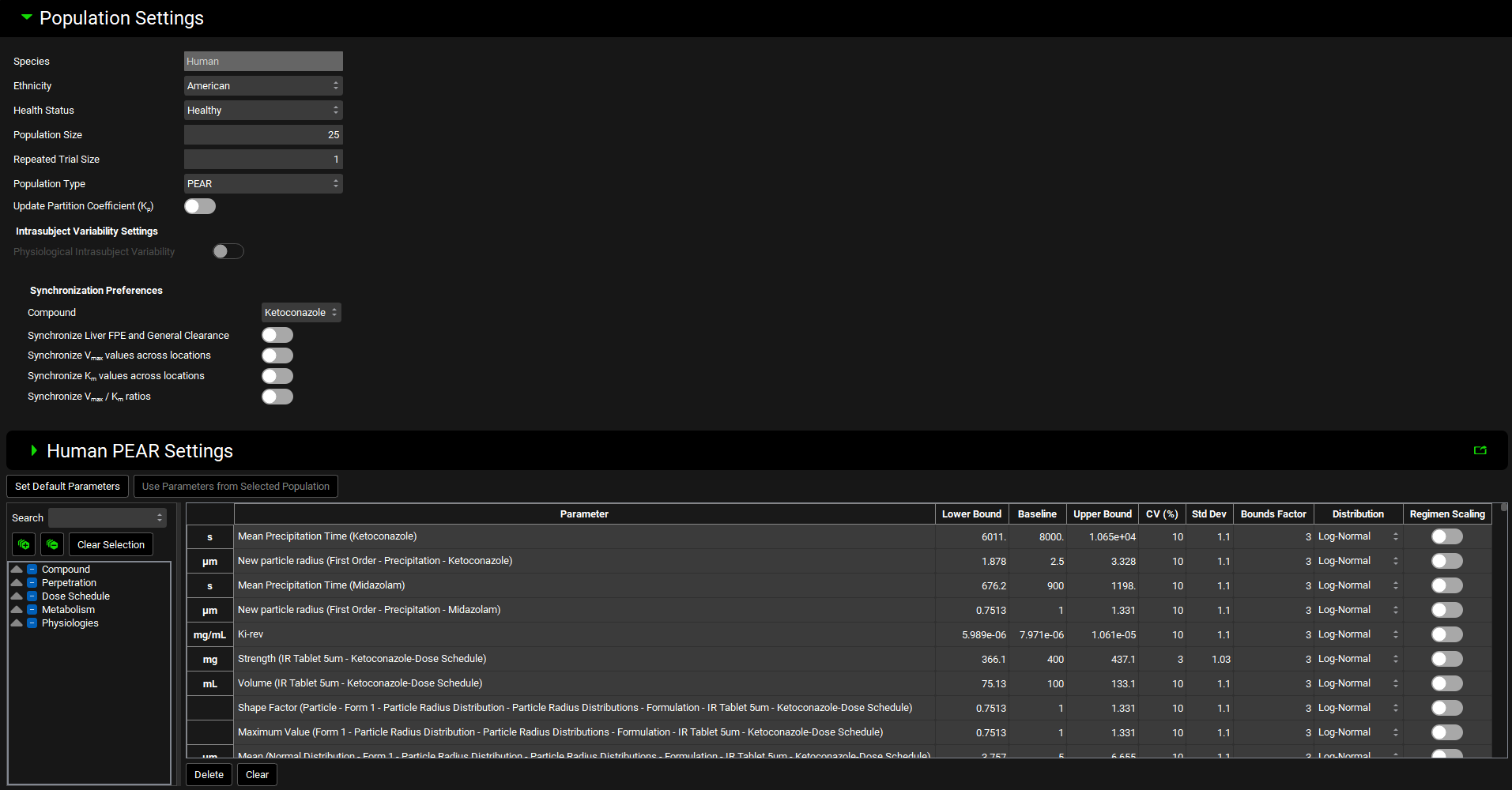
Population Settings panel
General population simulation settings are specified in this panel. The panel becomes enabled (1) when Dynamic Prediction Type and Population Simulation Mode are selected in the Configuration panel and (2) after the Victim and Perpetrator compounds have been selected in their respective Parent tables and relevant Perpetration Parameters have been selected.
Input/Option | Description |
Species | Simulations species (dependent on the Physiology Schedule selected in the Configuration panel). |
Ethnicity | Population to sample from (current options include American, Japanese, Chinese). |
Health Status | Health status for the generated virtual subjects (healthy, pregnant, different degrees of hepatic or renal impairment). |
Population Size | Number of virtual subjects to be created for each virtual population. |
Repeated Trial Size | Number of virtual populations to be created. |
Population Type | Parameter sampling strategy (PEAR – account for parameter covariates where applicable, applicable only with PBPK model, Monte Carlo – ignore parameter covariates and sample each parameter independently from its own distribution). |
Update Partition Coefficients (Kp) | A toggle. Turn on to update Kps for each virtual subject based on the parameter values selected for each subject and selected Kp method for the compound. |
Intrasubject Variability Settings | Section where intrasubject variability is set up. |
Physiological Intrasubject Variability | A toggle. Controls whether additional variability will be applied to gut physiological parameters (transit times, intestinal fluid volume, pH values, and bile salt concentrations for each gut section) in the same subject between simulations. |
Synchronization Preferences | Synchronize parameter values related to the compound and/or protein (enzyme or transporter).
|
Individual Parameter settings |
|
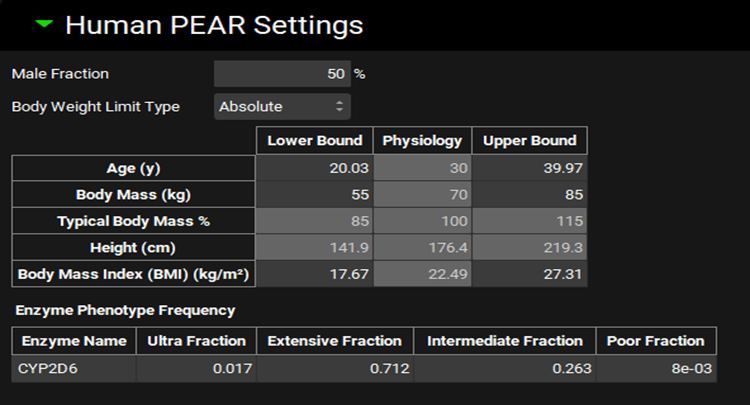
Human PEAR Settings sub-panel
When a PBPK model defined in the base simulations selected for the compounds is selected for the DDI prediction, the population specifications are defined in this sub-panel. The settings are not relevant if the simulations are using Compartmental PK models.
Input/Option | Description |
Male Fraction | % of males in the population. |
Body Weight Limit Type | Define the range of body weights for virtual subjects as absolute values in kg (Absolute) or as percent of the typical value for given subject (Percent). Percent selection is recommended for pediatric populations. |
Population Specification Table | Define range of demographic parameters (Age, Body Mass, or BMI) for virtual subjects included in the population simulation. Body Mass can be defined in kg or as % of typical body mass depending on selection in Body Weight Limit Type drop-down. Height is automatically calculated from Body Mass and BMI sampled for each virtual subject. Physiology column displays values from the physiology in the Physiology Schedule selected in the Configuration Panel. Enter the desired Lower Bound and Upper Bound for each parameter in their respective columns. |
Enzyme Phenotype Frequency | Define the proportion of subjects with different enzyme activities to be included in the virtual population. The table is enabled if the baseline model in the simulation includes enzymes with known polymorphism in their activity. |
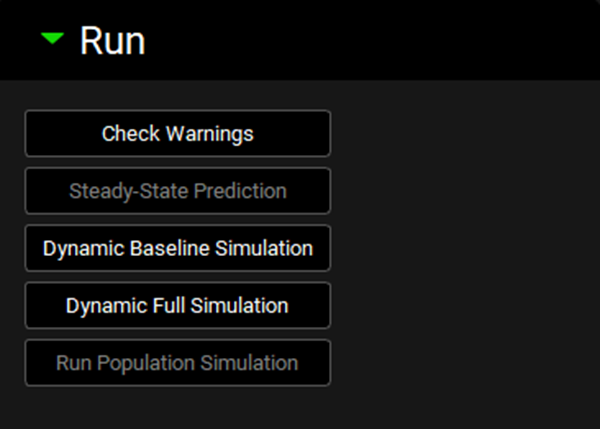
Run panel
This panel includes buttons to execute DDI predictions. Different buttons will be enabled depending on the Prediction type and Simulation Mode selected in the Configuration panel.
Input/Option | Description |
Check Warnings | Click this button to verify the DDI prediction settings and identify any errors that need to be addressed for accurate predictions. Any error messages will be displayed in the Message Center at the top of the interface. |
Steady-State Prediction | Predict DDI using Steady-State equation (enabled only when Steady State Prediction Type is selected in the Configuration panel). |
Dynamic Baseline Simulation | Complete a baseline simulation, i.e. simulation without any DDI impact (enabled only when Dynamic Single Simulation type is selected in the Configuration panel). |
Dynamic Full Simulation | Complete a full dynamic simulation, i.e. simulation accounting for DDIs based on the parameters selected in the Perpetration Parameters table (enabled only when Dynamic Single Simulation type is selected in the Configuration panel). |
Run Population Simulation | Complete a dynamic DDI prediction for a virtual population of subjects as defined in the Population Settings panel. The Population Simulation run automatically executes Baseline and Full simulation for each virtual subject. (enabled only when Dynamic Population Simulation type is selected in the Configuration panel). |
Results panel
Numerical predictions for all prediction types are displayed in this panel. If any simulations were completed as part of the prediction (i.e. with Dynamic prediction of if Simulated perpetrator concentrations were selected for Steady-State Prediction), the simulated profiles may be viewed in the Analysis view. We strongly recommend that you review those simulated profiles to verify the correct setup for the DDI prediction (i.e. to make sure that the dosing of perpetrator and victim compounds was correctly defined for DDI simulation as intended).
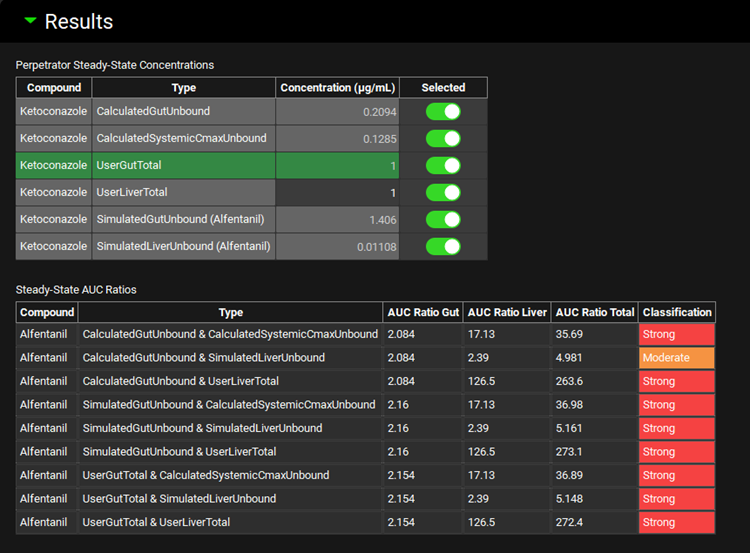
Steady State Predictions
Input/Option | Description |
Perpetrator Steady-State Concentrations | Summary of effective perpetrator concentrations selected in the Perpetrator Steady-State Concentrations sub-panel:
|
Steady-State AUC Ratios | Summary of AUC ratios predicted using the selected perpetrator concentrations.
|
Where units can be edited, this is done by clicking in the column header.
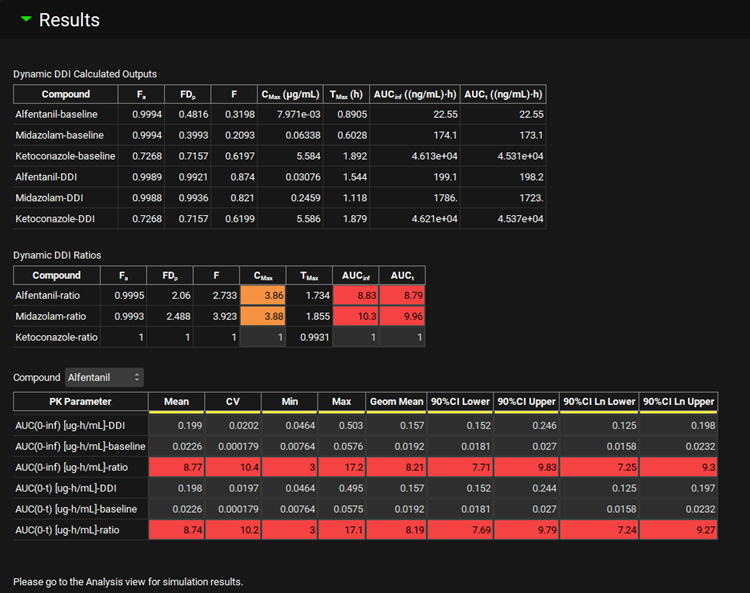
Dynamic Simulation Predictions
Input/Option | Description |
Dynamic DDI Calculated Outputs | Standard simulation endpoints from Baseline and Full Dynamic simulations for all compounds included in the dynamic DDI prediction:
In Population Simulation Mode, the values represent the average across all virtual subjects. Note: With multiple doses, the values represent the entire course of simulation. |
Dynamic DDI Ratios | Ratios of all simulation endpoints calculated as Full Dynamic Simulation/Baseline Simulation. In Population Simulation Mode, the values represent ratios of average values reported in the Dynamic DDI Calculated Outputs table. |
Compound | Select compound to display statistics for simulated PK endpoints. Displayed only with the Population Simulation Mode. |
Statistics Table | Summary statistics for simulated PK endpoints for compound selected in the drop-down above.
Displayed only with the Population Simulation Mode. |
Where units can be edited, this is done by clicking in the column header.
