Metabolism and Transport Module
The Metabolism and Transport module enables incorporation of saturable metabolism and carrier-mediated transport in the absorption and pharmacokinetic model. These processes are often parameterized based on in vitro experiments which produce results in a variety of different units. GastroPlus® includes a convenient units converter to enable quick conversion of in vitro metabolic or transport data to the in vivo values usable in the model. Specifically, experimental enzyme and transporter kinetics, metabolic clearance, and diffusional clearance measures may be converted to in vivo parameters, which can then be exported to the corresponding assets in the GPX™ project. This conversion is a form of in vitro to in vivo extrapolation (IVIVE) done via dimensional analysis, based on assay- and physiology- specific scaling factors.
The Metabolism and Transport Module provides access to the units converter and consists of a series of global settings at the top for selecting the compound, physiology, assay, and fraction unbound method. These are followed by three panels: Scaling Factors, Metabolism, and Transport. The selected assay will affect which panels are enabled, as well as certain values within.
The Metabolism and Transport module has three panels. All the inputs and options on these three panels (and where applicable, their sub-panels) are described in the following sections.
See:
Metabolism and Transport Module
Input/Option | Description |
Compound | A drop-down. Selects the compound for which the conversions will be conducted. The selection affects the numerical conversions (compound molecular weight is used in some conversions), and it will also affect where enzyme and transporter kinetic, as well as intrinsic clearance and PStc values, are exported. |
Physiology | A drop-down. Selects the physiology for which the conversions will be conducted. The selection here impacts both the conversions and where converted values will be exported. |
Assay | A drop-down. Selects the type of assay that was used to generate the in vitro data.
Enables the Transport panel below. |
In Vitro Fraction Unbound | A drop-down. Selects the method for calculating the fraction unbound in vitro (e.g. Fuinc, Fuhep, Fumic) in the assay. Options are dependent on the assay type:
|
Fraction | Displays the calculated fraction unbound in vitro, as a percentage, based on the method selected in the In Vitro Fraction Unbound drop-down. If the In Vitro Fraction Unbound method is set to User Defined, this field is editable and the user needs to specify the Fraction. For all other methods, this field is provided for information and is not editable. |
Scaling Factors panel
The Scaling Factors panel provides options for selecting the tissue type and weight for which the conversion will be conducted, as well as scaling factors for protein and/or cellular abundance. The options available, as well as the respective units, will vary based on the Assay selected above. Tissue, Tissue Weight Source, and Tissue Weight apply to all assay types; the remaining scaling factors will be assay specific.
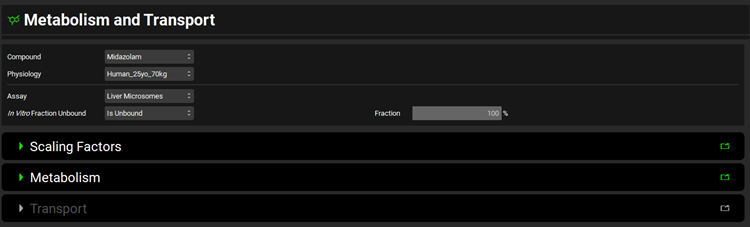
Metabolism and Transport Module, Scaling Factors panel (Liver Microsomes)
Input/Option | Description |
Tissue | A drop-down. Selects the tissue for which the conversions will be conducted. The selection here will affect the tissue weight and protein expression and will dictate the tissue to which converted intrinsic clearance and PStc values will be exported. The options available depend on the selection in Tissue Weight Source below. If Tissue Weight Source is set to Compartmental, only Liver will be available. If it is set to PBPK, all PEAR tissues will be available. |
Tissue Weight Source | A drop-down. Selects the source for the tissue weight: Compartmental or PBPK. If Compartmental is selected, the only tissue available will be the liver, and the default Tissue Weight will be set to 1500g. If PBPK is selected, all PEAR tissues in the Physiology selected at the top of the Metabolism and Transport module view will be available, and the Tissue Weight will correspond to the weight of the selected Tissue in that physiology. |
Tissue Weight | The mass, in g, of the selected tissue. The default value is based on the selection of Tissue and Tissue Weight Source above. The value generally should not be edited. Changing the Tissue or Tissue Weight Source selection above will reset the value. |
Microsomal Protein | The mass of microsomal protein, in mg, per the mass of tissue, in g. The value is based on the Tissue and the species and ethnicity (for humans) of the Physiology selected at the top of the Metabolism and Transport module view. This value is used in the calculation of the total protein expression in the tissue. The value can be edited. |
In Vitro Microsomal Protein Concentration
| The concentration, in mg/mL, or microsomal protein in the in vitro assay. This value is used in the calculation of In Vitro Fraction Unbound for the Austin and Hallifax methods. |
Restore Defaults | Restores any edited Scaling Factors to their default values. |
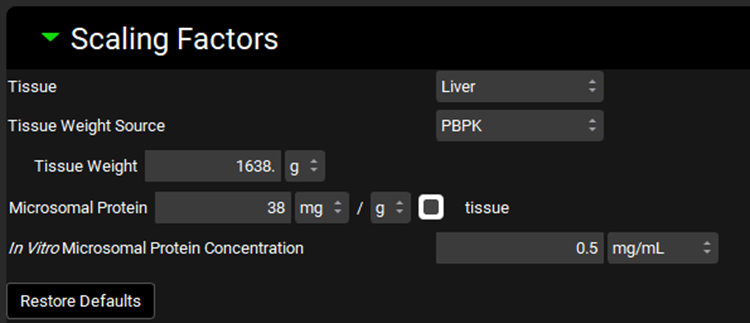
Metabolism and Transport Module, Scaling Factors panel (Hepatocytes)
Input/Option | Description |
Tissue | A drop-down. Selects the tissue for which the conversions will be conducted. The selection here will affect the tissue weight and protein expression and will dictate the tissue to which converted intrinsic clearance and PStc values will be exported. The options available depend on the selection in Tissue Weight Source below. If Tissue Weight Source is set to Compartmental, only Liver will be available. If it is set to PBPK, all PEAR tissues will be available. |
Tissue Weight Source | A drop-down. Selects the source for the tissue weight: Compartmental or PBPK. If Compartmental is selected, the only tissue available will be the liver, and the default Tissue Weight will be set to 1500g. If PBPK is selected, all PEAR tissues in the Physiology selected at the top of the Metabolism and Transport module view will be available, and the Tissue Weight will correspond to the weight of the selected Tissue in that physiology. |
Tissue Weight | The mass, in g, of the selected tissue. The default value is based on the selection of Tissue and Tissue Weight Source above. The value generally should not be edited. Changing the Tissue or Tissue Weight Source selection above will reset the value. |
Cells | The number of hepatocytes, in millions, per the mass of tissue, in g. The value is based on the Tissue and the species and ethnicity (for humans) of the Physiology selected at the top of the Metabolism and Transport module view. This value is used in the calculation of the total number of cells in the tissue. The value can be edited. |
Cellular Volume | The total volume, in mL, of the cells of the tissue. The value is based on the Tissue and the species and ethnicity (for humans) of the Physiology selected at the top of the Metabolism and Transport module view. This value is used in the calculation of Spec-PStc (see Transport panel below). The value generally should not be edited. |
In Vitro Hepatocyte Cellular Concentration
| The concentration, in million cells/mL, of hepatocytes in the in vitro assay. This value is used in the calculation of intrinsic clearance from half-life. The value can be edited. |
Expressed Protein | The mass of protein, in mg, expressed per the number of hepatocytes, in millions. This value is used in the calculation of the total protein expression in the tissue. The value can be edited. |
Restore Defaults | Restores any edited Scaling Factors to their default values. |
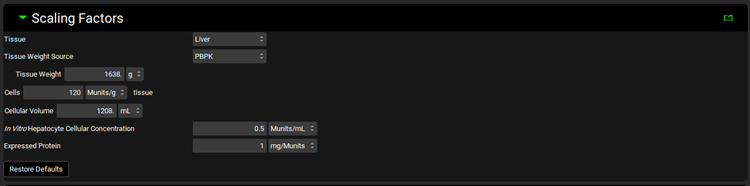
Metabolism and Transport Module, Scaling Factors panel (User Recombinant)
Input/Option | Description |
Tissue | A drop-down. Selects the tissue for which the conversions will be conducted. The selection here will affect the tissue weight and protein expression and will dictate the tissue to which converted intrinsic clearance and PStc values will be exported. The options available depend on the selection in Tissue Weight Source below. If Tissue Weight Source is set to Compartmental, only Liver will be available. If it is set to PBPK, all PEAR tissues will be available. |
Tissue Weight Source | A drop-down. Selects the source for the tissue weight: Compartmental or PBPK. If Compartmental is selected, the only tissue available will be the liver, and the default Tissue Weight will be set to 1500g. If PBPK is selected, all PEAR tissues in the Physiology selected at the top of the Metabolism and Transport module view will be available, and the Tissue Weight will correspond to the weight of the selected Tissue in that physiology. |
Tissue Weight | The mass, in g, of the selected tissue. The default value is based on the selection of Tissue and Tissue Weight Source above. The value can be edited. Changing the Tissue or Tissue Weight Source selection above will reset the value. |
Microsomal Protein | The mass of microsomal protein, in mg, per the mass of tissue, in g. The value is based on the Tissue and the species and ethnicity (for humans) of the Physiology selected at the top of the Metabolism and Transport module view. This value is used in the calculation of the total protein expression in the tissue. The value can be edited. |
In Vitro Recombinant Enzyme Protein Concentration
| The concentration, in mg/mL, of recombinant protein in the in vitro assay. This value is used in the calculation of In Vitro Fraction Unbound for the Austin and Hallifax methods. |
Restore Defaults | Restores any edited Scaling Factors to their default values. |
Metabolism and Transport Module, Scaling Factors panel (Cytosol)
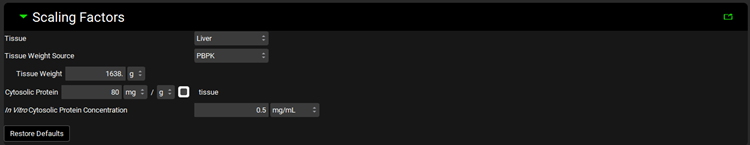
Input/Option | Description |
Tissue | A drop-down. Selects the tissue for which the conversions will be conducted. The selection here will affect the tissue weight and protein expression and will dictate the tissue to which converted intrinsic clearance and PStc values will be exported. The options available depend on the selection in Tissue Weight Source below. If Tissue Weight Source is set to Compartmental, only Liver will be available. If it is set to PBPK, all PEAR tissues will be available. |
Tissue Weight Source | A drop-down. Selects the source for the tissue weight: Compartmental or PBPK. If Compartmental is selected, the only tissue available will be the liver, and the default Tissue Weight will be set to 1500g. If PBPK is selected, all PEAR tissues in the Physiology selected at the top of the Metabolism and Transport module view will be available, and the Tissue Weight will correspond to the weight of the selected Tissue in that physiology. |
Tissue Weight | The mass, in g, of the selected tissue. The default value is based on the selection of Tissue and Tissue Weight Source above. The value generally should not be edited. Changing the Tissue or Tissue Weight Source selection above will reset the value. |
Cytosolic Protein | The mass of cytosolic protein, in mg, per the mass of tissue, in g. The value is based on the Tissue and the species and ethnicity (for humans) of the Physiology selected at the top of the Metabolism and Transport module view. This value is used in the calculation of the total protein expression in the tissue. The value can be edited. |
In Vitro Cytosolic Protein Concentration
| The concentration, in mg/mL, of cytosolic protein in the in vitro assay. This value is used in the calculation of In Vitro Fraction Unbound for the Austin and Hallifax methods. |
Restore Defaults | Restores any edited Scaling Factors to their default values. |
Metabolism and Transport Module, Scaling Factors panel (Expressed Cells)
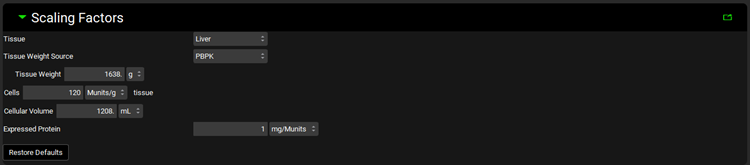
Input/Option | Description |
Tissue | A drop-down. Selects the tissue for which the conversions will be conducted. The selection here will affect the tissue weight and protein expression and will dictate the tissue to which converted intrinsic clearance and PStc values will be exported. The options available depend on the selection in Tissue Weight Source below. If Tissue Weight Source is set to Compartmental, only Liver will be available. If it is set to PBPK, all PEAR tissues will be available. |
Tissue Weight Source | A drop-down. Selects the source for the tissue weight: Compartmental or PBPK. If Compartmental is selected, the only tissue available will be the liver, and the default Tissue Weight will be set to 1500g. If PBPK is selected, all PEAR tissues in the Physiology selected at the top of the Metabolism and Transport module view will be available, and the Tissue Weight will correspond to the weight of the selected Tissue in that physiology. |
Tissue Weight | The mass, in g, of the selected tissue. The default value is based on the selection of Tissue and Tissue Weight Source above. The value generally should not be edited. Changing the Tissue or Tissue Weight Source selection above will reset the value. |
Cells | The number of cells, in millions, per the mass of tissue, in g. The value is based on the Tissue and the species and ethnicity (for humans) of the Physiology selected at the top of the Metabolism and Transport module view. This value is used in the calculation of the total protein expression in the tissue. The value can be edited. |
Cellular Volume | The total volume, in mL, of the cells of the tissue. The value is based on the Tissue and the species and ethnicity (for humans) of the Physiology selected at the top of the Metabolism and Transport module view. This value is used in the calculation of Spec-PStc (see Transport panel below). The value generally should not be edited. |
Expressed Protein | The mass of protein, in mg, expressed per the number of cells, in millions. This value is used in the calculation of the total protein expression in the tissue. The value can be edited. |
Restore Defaults | Restores any edited Scaling Factors to their default values. |
Metabolism panel
The Metabolism panel covers conversion of in vitro linear clearance measurements or Michaelis-Menten kinetics, handled by the Metabolic Clearance and Michaelis-Menten Kinetics sub-panels, respectively. In both cases, the in vitro data may be converted to either in vivo linear intrinsic clearance or in vivo Vmax and Km. The conversion between in vitro linear clearance and in vivo Michaelis-Menten kinetics is done under the assumption of non-saturated conditions. These conversions are based on the compound, physiology, assay, and fraction unbound method selected at the top of the view, as well as the enzyme selected in the Metabolism panel.
The Metabolism panel will be enabled only if the Assay type is set to Liver Microsomes, Hepatocytes, User Recombinant, or Cytosol. Additionally, at least one enzyme must be entered in the Enzyme Kinetics table for the compound selected at the top of the view (with a species that matches the species of the physiology selected at the top of the view) in order for Vmax, Km, and PBPK conversions on the sub-panels to be available.
The enzyme selection table at the top of the panel affects the conversion of in vitro Vmax and Km to both intrinsic clearance (Metabolic Clearance sub-panel) and in vivo Vmax and Km (Michaelis-Menten Kinetics sub-panel). The specific enzyme and phenotype selected will also determine to which enzyme converted in vivo Vmax and Km are exported.
Metabolism and Transport Module, Metabolism panel
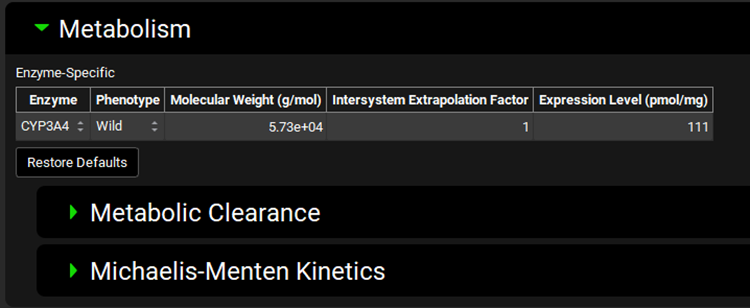
Input/Option | Description |
Enzyme | A drop-down. Selects the enzyme for which the conversion(s) will be done, as well as indicating for which enzyme the Molecular Weight, Intersystem Extrapolation Factor, and Expression Level will apply. The available enzymes are populated based on enzymes added to the enzyme kinetics table for the compound selected at the top of the view. The selection of enzyme here, combined with the phenotype selected, will control to which row(s) in the enzyme kinetics table the converted in vivo Vmax and Km will be exported. If no enzyme is selected here, Vmax, Km and PBPK conversions will not be functional in the sub-panels below. |
Phenotype | A drop-down. Selects the phenotype for the indicated enzyme. The selection here, combined with the selected enzyme, will control to which row(s) in the enzyme kinetics table the converted in vivo Vmax and Km will be exported. The selection of the phenotype will also impact the expression level (see below) for built-in enzymes for which phenotypic variations are known. |
Molecular Weight (g/mol) | The molecular weight, in g/mol, of the indicated enzyme. For built-in enzymes, this is populated automatically, and can be edited. For custom enzymes, this will need to be completed. |
Intersystem Extrapolation Factor | The Intersystem Extrapolation Factor (ISEF) for the enzyme. Represents in vivo:in vitro enzyme expression ratio and is applicable for conversion of in vitro data from User Recombinant Assay. Upon conversion the converted Vmax and intrinsic clearance values are multiplied by this factor. The default value is 1. The value can be edited. |
Expression Level | The expression level, in pmol-enzyme/mg-microsomal protein, of the indicated enzyme in the tissue selected in Scaling Factors panel. For built-in enzymes, this is populated automatically, and can be edited. For custom enzymes, this will need to be completed. |
Restore Defaults | Restores the Molecular Weight, Intersystem Extrapolation Factor, and Expression Level values for built-in enzymes to the default values. |
Metabolic Clearance sub-panel
The Metabolic Clearance sub-panel handles conversions of in vitro linear clearance measurements to either in vivo intrinsic clearance or in vivo non-saturated Vmax and Km.
Metabolism and Transport Module, Metabolism panel, Metabolic Clearance sub-panel (Source set to Clearance)
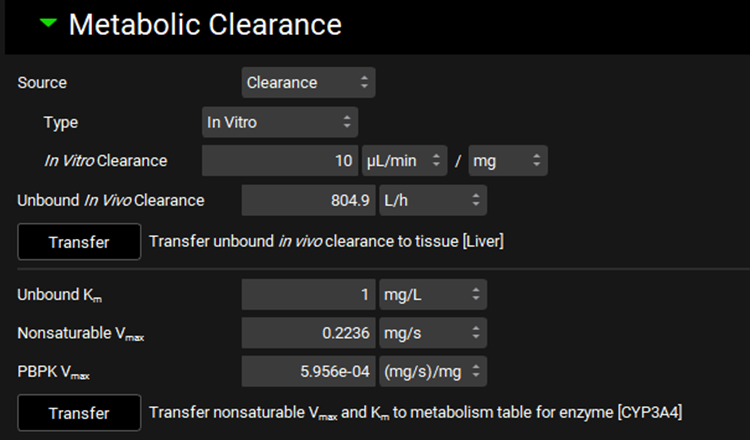
Metabolism and Transport Module, Metabolism panel, Metabolic Clearance sub-panel (Source set to HalfLife)
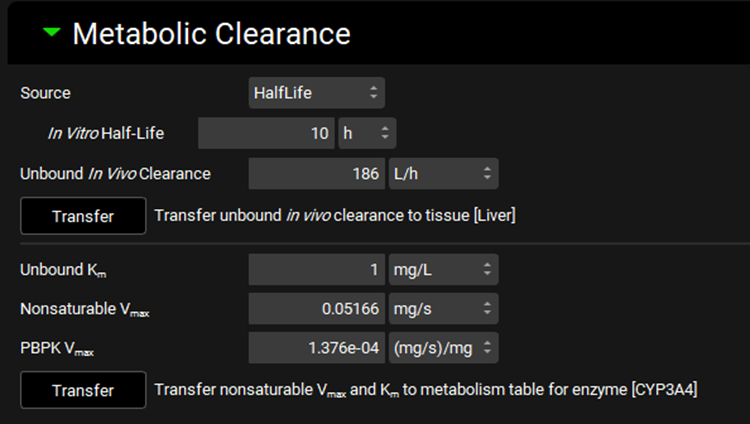
Input/Option | Description |
Source | A drop-down. Selects the measurement type: Clearance or Half-life.
|
Type | A drop-down. Selects the experiment type for in vitro experiments measuring clearance: In Vitro, Tissue, or Intrinsic. The specific options available depend on the Assay selected at the top of the view. The selection here will determine which units are available for the clearance entry field. Enabled only if Source is set to clearance. |
In Vitro Clearance | The measured in vitro clearance of the compound. The value can be edited. The options for units will be determined by the Type selected above. |
In Vitro Half-Life | The measured in vitro half-life of the compound. The value and the units (h, min, s) can be edited. |
Unbound In Vivo Clearance | The converted unbound in vivo clearance, in L/h. The value cannot be edited. |
Transfer | Transfers the Unbound In Vivo Clearance shown above to the tissue indicated to the right. The tissue is determined based on the Tissue selected in the Scaling Factors panel. This option is only available if the Tissue Weight Source in the Scaling Factors panel is set to PBPK. |
Unbound Km | The unbound in vivo Km, in mg/L, of the compound. This value is used in the calculation of Nonsaturable Vmax from in vitro linear clearance, using the assumption that CLint = Vmax / Km at non-saturated substrate concentrations. The value can be edited. |
Nonsaturable Vmax | The calculated in vivo Vmax, in mg/s, based on the converted Unbound In Vivo Clearance and the in vivo Unbound Km entered above. This calculation requires that either In Vitro Clearance or In Vitro Half-Life is entered above. Upon transfer (see below), this value will be applied to rows with Enterocyte, Lumen Dissolved, or Compartmental Liver locations in the Enzyme Kinetics table (see Enzyme Kinetics panel). The value cannot be edited. |
PBPK Vmax | The calculated in vivo PBPK Vmax, in mg/s/mg, based on the converted Unbound In Vivo Clearance and the in vivo Unbound Km entered above. This calculation requires that either In Vitro Clearance or In Vitro Half-Life is entered above. Upon transfer (see below), this value will be applied to rows with PBPK Tissue or Systemic Circulation locations in the Enzyme Kinetics table (see Enzyme Kinetics panel). The value cannot be edited. |
Transfer | Transfers the converted in vivo Nonsaturable Vmax and PBPK Vmax values, as well as the entered in vivo Km value, to the appropriate row(s) in the Enzyme Kinetics table for the compound selected at the top of the view. The enzyme is determined by the selection in the Enzyme Selection table above. |
Michaelis-Menten Kinetics sub-panel
The Michaelis-Menten Kinetics sub-panel handles conversions of in vitro Vmax and Km measurements to either in vivo Vmax and Km or in vivo intrinsic clearance.
Metabolism and Transport Module, Metabolism panel, Michaelis-Menten Kinetics sub-panel
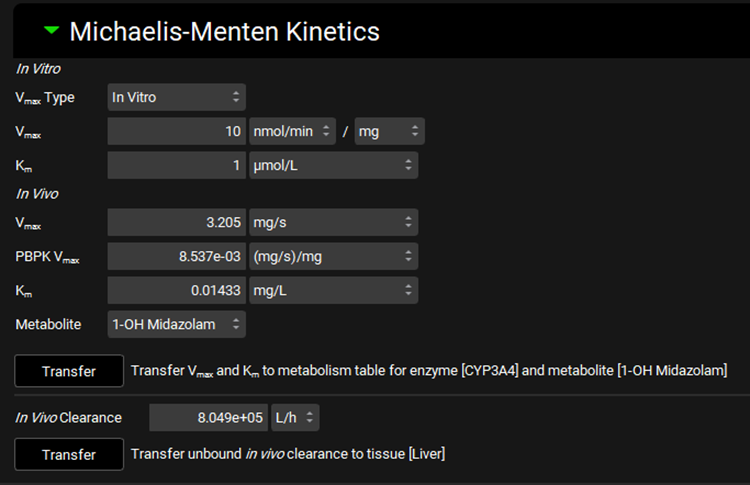
Input/Option | Description |
In Vitro | Area for the input of the measured in vitro values which will be converted. |
Vmax Type | A drop-down. Selects the experiment type for in vitro experiments measuring Michaelis-Menten kinetics: In Vitro or Intrinsic. The specific options available depend on the Assay selected at the top of the view. The selection here will determine which units are available for the Vmax entry field. |
Vmax | The measured in vitro Vmax value. The value can be edited. The options for units will be determined by the Vmax Type selected above. |
Km | The measured in vitro Km value. The value and the units (µmol/L, nmol/L, mmol/L) can be edited. |
In Vivo | Area which displays the converted in vivo values. |
Vmax | The calculated in vivo Vmax, in mg/s, based on the in vitro Vmax, assay setup, and scaling factors. This Vmax value corresponds to Enterocyte, Lumen Dissolved, and Compartmental Liver locations, with units of mg/s. The value cannot be edited. |
PBPK Vmax | The calculated in vivo PBPK Vmax, in mg/s/mg, based on the in vitro Vmax, assay setup, and scaling factors. This Vmax value corresponds to PBPK Tissue and Systemic Circulation locations, with units of mg/s/mg. The value cannot be edited. |
Km | The calculated unbound in vivo Km, in mg/L, based on the in vitro Km and fraction unbound in vitro. |
Metabolite | A drop-down. Selects the metabolite, if one has been specified, for the metabolism. The options are populated based on metabolites specified in the Enzyme Kinetics table (see Enzyme Kinetics panel) for the enzyme and phenotype selected in the Enzyme Selection table in the Metabolism panel above. If no metabolites have been specified, the option ‘None’ will be available and selected by default. The selection here determines to which row(s) in the Enzyme Kinetics table the converted Vmax and Km values will be exported upon Transfer. This does not affect the conversion itself. |
Transfer | Transfers the converted in vivo Vmax, PBPK Vmax, and Km values to the appropriate row(s) in the Enzyme Kinetics table for the compound selected at the top of the view. The enzyme is determined by the selection in the Enzyme Selection table above. |
In Vitro Clearance | The converted unbound in vivo clearance of the compound, based on the entered in vitro Vmax and Km values, under the assumption that CLint = Vmax / Km at non-saturated substrate concentrations. |
Transfer | Transfers the In Vivo Clearance shown above to the tissue indicated to the right. The tissue is determined based on the Tissue selected in the Scaling Factors panel. This option is only available if the Tissue Weight Source in the Scaling Factors panel is set to PBPK. |
Transport panel
The Transport panel covers conversion of in vitro Michaelis-Menten transporter kinetics or diffusional clearance measurements to in vivo transporter kinetics or PStc values, respectively. These two types of conversions are handled separately.
The Transport panel will be enabled only if the Assay type is set to Hepatocytes or Expressed Cells. Additionally, at least one transporter must be entered in the Transporter Kinetics table for the compound selected at the top of the view (with a species that matches the species of the physiology selected at the top of the view) in order for the transport conversions on this panel to be available. Conversion for diffusional clearance to PStc are available regardless of transporter entry.
The transporter selection table at the top of the panel affects the conversion of in vitro Vmax and Km to in vivo Vmax and Km. The specific transporter selected will also determine to which transporter converted in vivo Vmax and Km are exported.
Metabolism and Transport Module, Transport panel
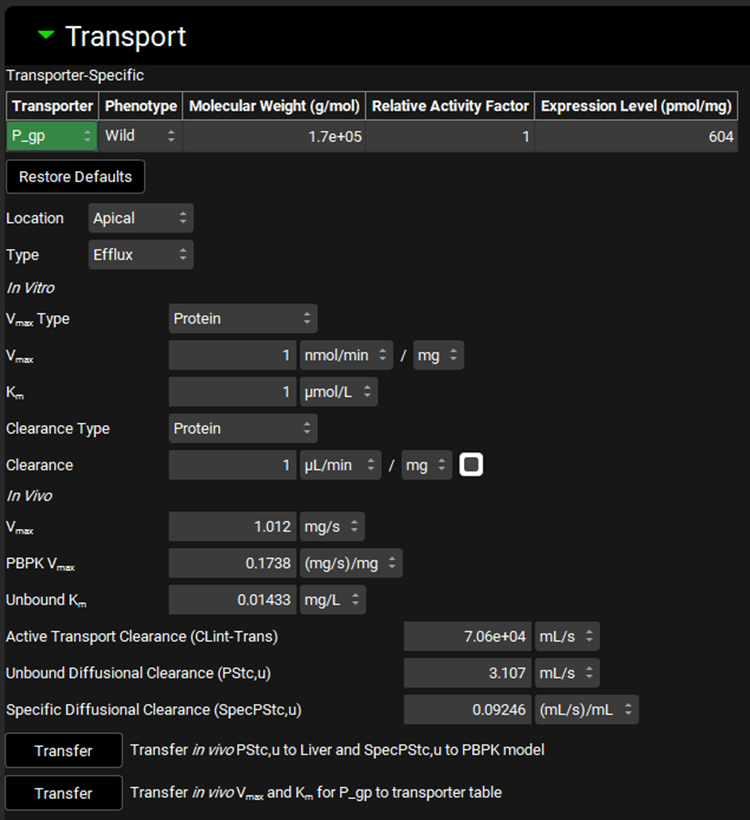
Input/Option | Description |
Transporter | A drop-down. Selects the transporter for which the conversion(s) will be done, as well as indicating for which transporter the Molecular Weight, Relative Activity Factor, and Expression Level will apply. The available transporters are populated based on the transporters added to the Transporter Kinetics table for the compound selected at the top of the view. The selection of transporter here, combined with the phenotype selected, will control to which row(s) in the transporter kinetics table the converted in vivo Vmax & Km will be exported. The selection here has no impact on diffusional clearance conversions. |
Phenotype | A drop-down. Selects the phenotype for the indicated transporter. |
Molecular Weight (g/mol) | The molecular weight, in g/mol, of the indicated transporter. For built-in transporters, this is populated automatically, and can be edited. For custom transporters, this will need to be completed. |
Relative Activity Factor | The Relative Activity Factor for the transporter. Represents in vivo:in vitro transporter activity ratio. Upon conversion the converted Vmax values are multiplied by this factor. The default value is 1. The value can be edited. |
Expression Level | The expression level, in pmol- transporter/mg-expressed protein, of the indicated transporter in the selected tissue. For built-in transporters, this is populated automatically, and can be edited. For custom transporters, this will need to be completed. |
Restore Defaults | Restores the Molecular Weight, Relative Activity Factor, and Expression Level values for built-in transporters to the default values. |
Location | A drop-down. Selects the membrane location for flux: Apical or Basolateral. The available options are based on the Tissue selected in the Scaling Factors panel above. Basolateral is available for all tissues, while Apical is available only for liver, lung, and kidney. The selection affects to which side of the tissue, apical or basolateral, calculated PStc values will be exported (see Compound sub panel - Tissues). |
Type | A drop-down. Selects the type of flux for active transport: Influx or Efflux. |
In Vitro | Area for the input of the measured in vitro values which will be converted. |
Vmax Type | A drop-down. Selects the experiment type for in vitro experiments measuring Michaelis-Menten kinetics: Protein, Cells, or Intrinsic. The selection here will determine which units are available for the Vmax entry field. |
Vmax | The measured in vitro Vmax value. The value can be edited. The options for units will be determined by the Vmax Type selected above. |
Km | The measured in vitro Km value. The value and the units (µmol/L, nmol/L, mmol/L) can be edited. |
Clearance Type | A drop-down. Selects the experiment type for in vitro experiments measuring diffusional clearance: Protein, Tissue, Cells, or Intrinsic. The selection here will determine which units are available for the Clearance entry field. |
Clearance | The measured in vitro diffusional clearance value. |
In Vivo | Area which displays the converted in vivo values. |
Vmax | The calculated in vivo Vmax, in mg/s, based on the in vitro Vmax, assay setup, and scaling factors. This Vmax value corresponds to the Enterocyte location, with units of mg/s. The value cannot be edited. |
PBPK Vmax | The calculated in vivo PBPK Vmax, in mg/s/mg, based on the in vitro Vmax, assay setup, and scaling factors. This Vmax value corresponds to the PBPK Tissue location, with units of mg/s/mg. The value cannot be edited. |
Unbound Km | The calculated unbound in vivo Km, in mg/L, based on the in vitro Km and fraction unbound in vitro. |
Active Transport Clearance (CLint-Trans) | The calculated in vivo intrinsic clearance, in mL/s, of the transporter, based on the converted in vivo Vmax and Km, using the equation CLint = Vmax / Km. This value is provided for informational purposes only and is not used in the model. |
Unbound Diffusional Clearance (PStc,u) | The calculated in vivo diffusional clearance (PStc) value, in mL/s, based on the in vitro diffusional clearance, assay setup, scaling factors, and fraction unbound in vitro. The value cannot be edited. |
Specific Diffusional Clearance (SpecPStc,u) | The calculated in vivo specific diffusional clearance (SpecPStc) value, in (mL/s)/mL. This value is calculated from Unbound Diffusional Clearance (PStc,u), Cellular Volume (see Scaling Factors panel), land Location. The value cannot be edited. |
Transfer (PStc,u and SpecPStc,u) | Transfers the converted In Vivo Unbound Diffusional Clearance (PStc,u) shown above to the tissue indicated to the right. The tissue is determined based on the Tissue selected in the Scaling Factors panel. The selection made in the Location drop-down above will determine whether this is transferred as the apical or basolateral PStc for the indicated tissue. Also transfers the converted In Vivo Specific Unbound Diffusional Clearance (SpecPStc,u) shown above to the PBPK model linked to the compound and physiology selected at the top of the Module. The option to use SpecPStc can then be toggled on within the PBPK panel (see PBPKPlus™ Module). This option is only available if the Tissue Weight Source in the Scaling Factors panel is set to PBPK. |
Transfer (Vmax and Km) | Transfers the converted in vivo Vmax, PBPK Vmax, and Km values to the appropriate row(s) in the Transporter Kinetics table for the compound selected at the top of the view. The transporter is determined by the selection in the Transporter Selection table above. |
